
Biologic therapeutics such as monoclonal antibodies and biosimilars are complex proteins that are susceptible to post-translational modifications (PTMs). These chemical modifications can affect the performance and activity of the biologic, potentially resulting in decreased potency and increased immunogenicity. Such modifications include glycosylation, deamidation, oxidation and disulfide bond shuffling. These PTMs can be signs of protein degradation, manufacturing issues or improper storage. Several of these modifications are well characterized, and methods exist for detecting them during biologic manufacture. However, disulfide shuffling is not particularly well characterized for biologics, and no methods exist to easily detect and quantify disulfide bond shuffling in biologics.
Normally the cysteines in a protein will pair with a predictable or “normal” partner residue either within a polypeptide chain or between two polypeptide chains when they form disulfide bonds. These normal disulfide bonds are important for final protein conformation and stability. Indeed, disulfide bonds are considered an important quality indicator for biologics.
In a recently published study, Coghlan and colleagues designed a semi-automated method for characterizing disulfide bond shuffling on two IgG1 biologics: rituximab (originator drug Rituxan® and biosimilar Acellbia®) and bevacizumab (originator Avastin® and biosimilar Avegra®).
Disulfide Bond Shuffling in IgG1 Proteins
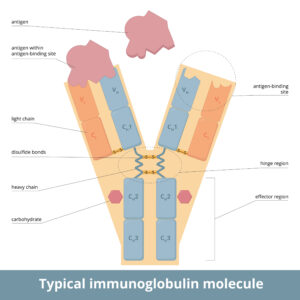
In the study reviewed here, Coghlan and colleagues compare rituximab or bevacizumab originators and biosimilars. These biologics are IgG1 proteins that typically contain 16 predicted disulfide bonds. Four of them are interchain, occurring between the two heavy chains of the constant region of the antibody, and one each between the constant regions of the heavy and light chains near the hinge. The remaining 12 disulfide bonds occur throughout the constant and variable regions of the antibody on both the heavy and light chains.
Disulfide bond shuffling (or scrambling) occurs when an S-S bond between a pair of cysteine residues is rearranged such that the residues are pairing with unpredicted partners. Often disulfide bond shuffling is observed when a protein is exposed to adverse conditions such as heat, free radicals, high pH or agitation. The four interchain disulfide bonds of IgG1s tend to be more susceptible to reduction, which makes them more susceptible to disulfide bond shuffling.
Are you looking for proteases to use in your research?
Explore our portfolio of proteases today.
In the method described by Coghlan et al., antibodies were denatured and free cysteines blocked prior to protein digestion conducted under low pH conditions. They used the Agilent AssayMAP Bravo liquid handling platform with the AccuMAP™ Low pH Protein Digestion Kit (AccuMAP denaturing solution, 10X low pH AccuMAP reaction buffer, AccuMAP low pH resistant rLys-C, N-ethylmaleimide (NEM)) and Trypsin Platinum. Samples were analyzed by tandem liquid chromatography-mass spectrometry.
The semi-automated mass spec analysis, combined with more traditional digestion and SDS-PAGE experiments, enabled Coghlan et al. to determine how bevacizumab and rituximab are different based on disulfide bond profile and protein degradation patterns. The authors detected disulfide bond shuffling in both antibodies, but how the proteins changed over time was different.
Importantly, the semi-automated method the researchers developed enabled them to better understand observed differences in the degradation and aggregation characteristics of drugs of the same IgG subclass. They suggest the semi-automated LC-MS/MS method developed in this study, along with SEC and SDS-PAGE analysis, allows drug developers to identify, count and characterize disulfide bond formation during biologic drug development. Because the process is automated, variability in workflows is reduced, providing the groundwork for standardized methods to detect and characterize protein degradation.
Literature Cited
Coghlan, J. et al. (2022) Streamlining the characterization of disulfide bond shuffling and protein degradation in IgG1 biopharmaceuticals under native and stressed conditions. Front. Bioeng. Biotechnol. 10: 862456 [doi: 10.3389/fbioe.2022.862456]
Like what you read? Try a sample of high-efficiency Trypsin Platinum today!
Related Posts
Michele Arduengo
Latest posts by Michele Arduengo (see all)
- IL-6/STAT3-Regulated Long Non-Coding RNA Is Involved in Colorectal Cancer Progression - July 1, 2025
- Using Dual-Luciferase Assays to Identify the Role of Non-Coding RNAs in Disease - May 2, 2025
- An Unexpected Role for RNA Methylation in Mitosis Leads to New Understanding of Neurodevelopmental Disorders - March 27, 2025
