This blog was written by Sebastien Smick, Research Technician in Dr. Jacquin Niles’ laboratory at Massachusetts Institute of Technology (MIT)
Our lab is heavily focused on the basic biology and drug discovery of the human bloodborne pathogen Plasmodium falciparum, which causes malaria. We use the CRISPR/Cas9 system, paired with a TetR protein fused to a native translational repressor alongside a Renilla luciferase reporter gene, to conditionally knock down genes of interest to create modified parasites. We can then test all kinds of compounds as potential drug scaffolds against these gene-edited parasites. Our most recent endeavor, one made possible by Promega, is a medium-low throughput robotic screening pipeline which compares conditionally-activated or-repressed parasites against our dose-response drug libraries in a 384-well format. This process has been developed over the past few years and is a major upgrade for our lab in terms of data production. Our researchers are working very hard to generate new modified parasites to test. Our robots and plate readers rarely get a day’s rest!
Bioluminescence
Toxicity Studies in Organoid Models: Developing an Alternative to Animal Testing
Alternatives to animal testing have long been explored when it comes to studying the safety of various chemical compounds for use in food, medicine and cosmetics. With the advent of three-dimensional (3D) cell culture to create organoids, more relevant human organoid models are being explored as one way to provide safe and effective compound testing while minimizing the need for testing in animals. The international project Physiologically Anchored Tools for Realistic nanOmateriaL hazard aSsessment (PATROLS) led by the Swansea University Medical School aims to establish a battery of innovative, next-generation safety testing tools that can more accurately predict the effects of engineered nanomaterial (ENM) exposure in humans and the environment.
One project being researched by Samantha Llewellyn, a research assistant at Swansea University, is developing predictive physiologically relevant 3D liver models for ENM safety assessment. By having a model to evaluate realistic ENM exposures, a researcher can study liver function, hepatic metabolism and microtissue cell viability after acute (24 hours) or prolonged (several days) exposure. A microtissue model for assessing ENM hepatotoxicity needs to mimic primary hepatocytes and be amenable to assays used to test cell viability and metabolism.
The right tools for testing this 3D liver model include the bioluminescent-based CellTiter-Glo® 3D Viability and P450-Glo® Assays. When creating organoids, having reagents that can penetrate to the center of the dense and complex 3D liver spheroids is important so that the cell viability readout encompasses the entire microtissue. The CellTiter-Glo® 3D Viability Assay accomplishes this task, providing accurate assessment of 3D tissue cell health. Measuring cytochrome P450 (CYP450) activity is necessary for studying liver function. The P450-Glo® Assays have the flexibility to assess CYP450 activity while preserving the liver spheroids; thus, researchers can gather more data from a single experiment.
The importance of Samantha Llewellyn’s research as part of PATROLs is establishing a 3D liver model that could evaluate realistic ENM exposures and reduce the need for animal testing. Bioluminescent assays for assessing cell health and liver enzyme function are necessary to reach this goal.

To learn more about the last 30 years of bioluminescent innovations and the discoveries they’ve enabled, please visit our 30th anniversary celebration page.
Related Posts
RNA-Protein Interactions: A New Frontier for Drug Discovery
Almost 90% of the human genome is transcribed into RNA, but only 3% is ultimately translated into a protein. Some non-translated RNA is thought to be useless, while some play a significant yet often mysterious role in cancer and other diseases. Despite its abundance and biological significance, RNA is rarely the target of therapeutics.
“We say it’s undruggable, but I would say that ‘not-yet-drugged’ is a better way to put it,” says Amanda Garner, Associate Professor of Medicinal Chemistry at the University of Michigan. “We know that RNA biology is important, but we don’t yet know how to target it.”

Amanda’s lab develops systems to study RNA biology. She employs a variety of approaches to analyze the functions of different RNAs and study their interactions with proteins. Her lab recently published a paper describing a novel method for studying RNA-protein interactions (RPI) in live cells. Amanda says that with the right tools, RPI could become a critical target for drug discovery.
“It’s amazing that current drugs ever work, because they’re all based on really old approaches,” Amanda says. “This isn’t going to be like developing a small molecule kinase inhibitor. It’s a whole new world.”
Continue reading “RNA-Protein Interactions: A New Frontier for Drug Discovery”Drug Repurposing Screens: Redeploying Old Dogs for New Tricks
This blog was written by guest author, Amy Landreman, PhD.
Drug repurposing, identifying new uses for approved or investigational drugs, is an attractive strategy when looking for new disease treatments. Because the compounds have already gone through some level of pre-clinical optimization and safety testing, this approach can reduce risk, reduce cost, and speed up the timeline for further drug development. An additional benefit of this approach is that it can result in new biological insights or a better understanding of disease mechanisms since these compounds usually already have some level of mechanistic characterization. Indeed, there are now a number of compound collections openly available specifically for the purpose of facilitating drug repurposing efforts. For example, the ReFRAME (Repurposing, Focused Rescue, and Accelerated Medchem) library is a collection of 12,000 compounds developed by Scripps Research Center and has been screened to identify novel candidate therapeutics for Cryptosporidium infection (1). The Broad Institute also offers a drug repurposing hub that contains an annotated collection of over 7,000 compounds.

Drug repurposing libraries, although often smaller than novel compound small molecule libraries, are designed for implementation into high-throughput screening workflows in order to efficiently triage compounds for the desired result. Effective compound screens require assays that can be scaled to 384 or 1536-well microplate formats and implemented in batch or continuous processing workflows. The firefly luciferase reaction has been leveraged to create many assays that are well-suited to these types of high-throughput screening approaches. In particular, the generation of “Glow” assays that have stable luminescent signals and homogenous assay design is a good fit. The signal stability allows for multi-plate processing and because the reagent is added directly to cells in culture, pre-processing steps are eliminated allowing for automated workflows. Assay reagents such as the CellTiter-Glo® Cell Viability Assay and the ADP-Glo™ Kinase Assay are commonly used in screening efforts including those done with repurposing libraries. In addition, there are several firefly luciferase reporter assay reagents such as Steady-Glo® and Bright-Glo™ Luciferase Assays that have been optimized for high-throughput detection of firefly luciferase activity making them well-suited to repurposing screens.
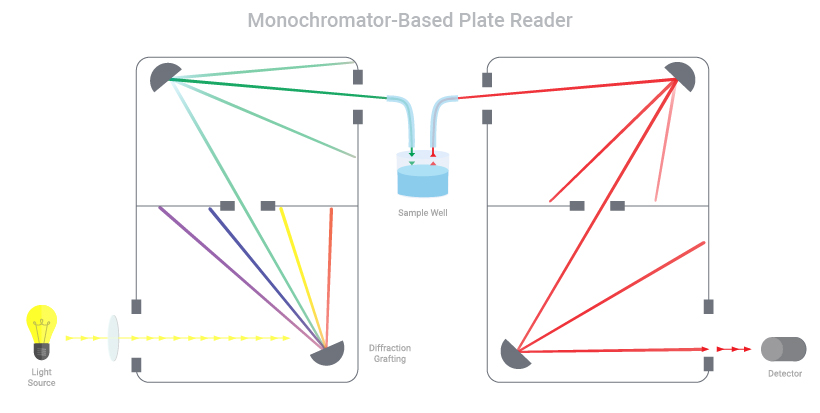
Monochromator vs Filter-Based Plate Reader: Which is Better?
When it comes to purchasing a microplate reader for fluorescence detection, the most common question is whether to choose a monochromator-based reader or filter-based reader. In this blog, we’ll discuss how both types of plate readers work and factors to consider when determining the best plate reader for your need.
How do monochromator-based plate readers work?
Monochromators work by taking a light source and splitting the light to focus a particular wavelength on the sample. During excitation, the light passes through a narrow slit, directed by a series of mirrors and diffraction grating and then passes through a second narrow slit prior to reaching the sample. This ensures the desired wavelength is selected to excite the fluorophore. Once the fluorophore is excited, it emits light at a different, longer wavelength. This emission light is captured by another series of mirrors, grating and slits to limit the emission to a desired wavelength, which then enters a detector for signal readout.
A SARS-CoV-2 NanoLuc® Reporter Virus for Rapid Screening of Antivirals
Before the COVID-19 global pandemic began, Dr. Xuping Xie, Assistant Professor of the University of Texas Medical Branch at Galveston, TX has been studying viruses, such as Dengue and Zika, for more than 10 years. Once the pandemic hit in early 2020, he was prepared to join the fight against the virus. “There was an urgent need to know: Is there a quicker way to develop therapeutics or antibodies to target SARS-CoV-2?” says Dr. Xie. “That’s why we immediately launched our SARS-CoV-2 project.”
His goal was to create an assay that could 1) screen for antiviral drugs and 2) quickly measure neutralizing antibody levels. The assay could be used to determine the immune status of previously infected individuals and to evaluate various vaccines under development. To achieve this, he wanted to create a reporter virus that is genetically stable and replicates similarly to the wild-type virus in cell culture.
Continue reading “A SARS-CoV-2 NanoLuc® Reporter Virus for Rapid Screening of Antivirals”Firefly Luciferase Sheds Light on Development of New Malaria Treatments

Despite significant advancements in antimalarial drugs and widespread efforts to prevent transmission over the past decade, deaths from malaria remain high, particularly in younger children. New drugs with novel modes of action are urgently needed to continue reducing mortality and address drug resistance in the malaria parasite, Plasmodium falciparum. While tens of thousands of compounds have been identified as potential candidates through massive screening efforts, scalable methods for identifying the most effective compounds are needed.
The goal is to find a drug that is potent during all stages in the life cycle of P. falciparum and kills the parasite quickly. Focusing on assessing whether a compound can rapidly eliminate initial parasite burden, Paul Horrocks, PhD, and his colleagues developed a validated bioluminescence-based assay that rapidly determines the initial rate of kill for discovery antimalarials. One key to developing their assay was figuring out how to monitor when the parasite dies after introducing the drug. While measuring DNA content can be used to monitor parasite burden, it is too stable to use for a relevant time course assay.
Enter firefly luciferase, a dynamic reporter tool to investigate drug action. By creating transgenic P. falciparum that express the luc reporter gene, the researchers could monitor drug action over time. When the parasite is killed, it stops making the luciferase reporter. Since there is no new production of luciferase, levels fall quickly after the parasite dies, and a luciferase assay can determine how fast each drug killed the parasite.
Continue reading “Firefly Luciferase Sheds Light on Development of New Malaria Treatments”Bioluminescent Sharks Set the Sea Aglow
Many deep sea creatures are bioluminescent. However, before documenting the luminescence of the kitefin shark, Dalatias licha, there has never been a nearly six-foot long luminous vertebrate creature. In a recent study, Mallefet and colleagues examined three species of sharks: Dalatias licha, Etmopterous lucifer, and Emopterus granulosus and documented their luminescence for the first time. These bioluminescent sharks are the largest bioluminescent creatures known.

Advancing Understanding of Hypoxic Gene Regulation Using Reporter Genes: Celebrating the Work of Dr. Gregg L. Semenza
This post is written by guest blogger, Amy Landreman, PhD, Sr. Product Manager at Promega Corporation.
Oxygen is necessary for animal life. It’s essential for cellular respiration and the production of energy (ATP) we require to survive. Given the need for oxygen, it isn’t surprising that our bodies have evolved ways to sense and adapt to decreased oxygen conditions (hypoxia). We can increase the production of new blood vessels by producing vascular endothelial growth factor (VEGF) or increase red blood cell (RBC) production by increasing the levels of eythropoietin (EPO), the hormone that plays a key role in the production of RBCs. But how does our body sense low oxygen, increase EPO levels, and kick our RBC production into gear? Nobel laureate Gregg L. Semenza has been honored for his contributions to our understanding of this process, and his research demonstrates the value of reporter genes and bioluminescence for studying gene regulation.
A Bioluminescent Biosensor for Detection of Mycotoxins in Food
Food contamination is a serious global health issue. According to the WHO, an estimated 600 million, almost 1 in 10 people globally, suffer from illness after eating contaminated food—and 420,000 die. Developing new technologies for more effective testing of food contaminants can help reduce that number and improve public health.
A recent application of bioluminescent technology could change the way we test for mycotoxins in the future. Dr. Jae-Hyuk Yu, Professor of Bacteriology at the University of Wisconsin-Madison, and his then graduate student, Dr. Tawfiq Alsulami, collaborated with Promega to develop a bioluminescent biosensor that enables simple and rapid detection of mycotoxins in food samples.
Continue reading “A Bioluminescent Biosensor for Detection of Mycotoxins in Food”