N-Glycosylation is a common protein post-translational modification occurring on asparagine residues of the consensus sequence asparagine-X-serine/threonine, where X may be any amino acid except proline. Protein N-glycosylation takes place in the endoplasmic reticulum (ER) as well as in the Golgi apparatus.
Approximately half of all proteins typically expressed in a cell undergo this modification, which entails the covalent addition of sugar moieties to specific amino acids. There are many potential functions of glycosylation. For instance, physical properties include: folding, trafficking, packing, stabilization and protease protection. N-glycans present at the cell surface are directly involved in cell−cell or cell−protein interactions that trigger various biological responses.
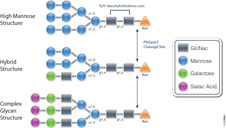
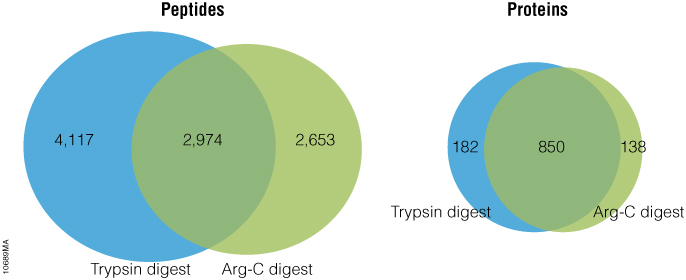
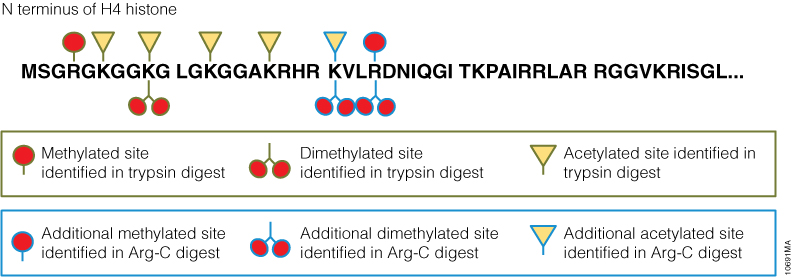
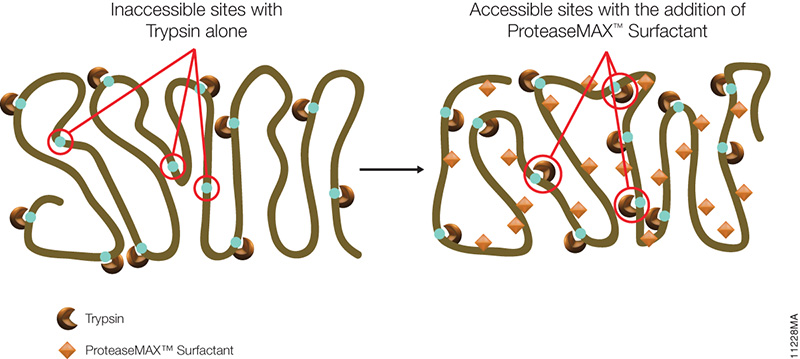
The standard method used to profile the N-glycosylation pattern of cells is glycoprotein isolation followed by denaturation and/or tryptic digestion of the glycoproteins and an enzymatic release of the N-glycans using PNGase F followed by analysis mass spec. This method has been reported to yield high levels of high-mannose N-glycans that stem from both membrane proteins as well as proteins from the ER.(1,2)
Are you looking for proteases to use in your research?
Explore our portfolio of proteases today.
For those researchers interested in characterizing only cell surface glycans (i.e., complex N-glycans) a recent reference has developed a model system using HEK-292 cells that demonstrates a reproducible, sensitive, and fast method to profile surface N-glycosylation from living cells (3). The method involves standard centrifugation followed by enzymatic release of cell surface N-glycans. When compared to the standard methods the detection and quantification of complex-type N-glycans by increased their relative amount from 14 to 85%.
- North, S. J. et al. (2012) Glycomic analysis of human mast cells, eosinophils and basophils. Glycobiology. 2012, 22, 12–22.
- Reinke, S. O. et al. (2011) Analysis of cell surface N-glycosylation of the human embryonic
kidney 293T cell line. J. Carbohydr. Chem. 30, 218–232. - Hamouda, H. et al. (2014) Rapid Analysis of Cell Surface N‑Glycosylation from Living Cells Using Mass Spectrometry. J of Proteome Res. 13, 6144–51.
 Neonatal sepsis is a systemic infection prevalent in preterm and very low birth weight infants and causes high morbidity. Most cases of neonatal sepsis are caused by pathogenic bacteria that invade the bloodstream, triggering an abrupt and overwhelming infection in the target organs accompanied by a systemic inflammatory response. Testing for neonatal sepsis is challenging because it does not affect a specific organ and presents multiple symptoms that are often confused with other related conditions (1). Current diagnostic tests for sepsis include those that identify markers of the host response to infection (e.g., procalcitonin, C reactive protein, cytokines, etc.) and those that detect bacterial infection in blood (bacteremia) (2). The lack of specific diagnostic biomarkers for early and accurate detection of neonatal sepsis has spurred the quest for next-generation biomarkers using powerful mass screening technologies such as proteomics.
Neonatal sepsis is a systemic infection prevalent in preterm and very low birth weight infants and causes high morbidity. Most cases of neonatal sepsis are caused by pathogenic bacteria that invade the bloodstream, triggering an abrupt and overwhelming infection in the target organs accompanied by a systemic inflammatory response. Testing for neonatal sepsis is challenging because it does not affect a specific organ and presents multiple symptoms that are often confused with other related conditions (1). Current diagnostic tests for sepsis include those that identify markers of the host response to infection (e.g., procalcitonin, C reactive protein, cytokines, etc.) and those that detect bacterial infection in blood (bacteremia) (2). The lack of specific diagnostic biomarkers for early and accurate detection of neonatal sepsis has spurred the quest for next-generation biomarkers using powerful mass screening technologies such as proteomics.