We have all been hearing a lot about RT-PCR, rRT-PCR and RT-qPCR lately, and for good reason. Real-Time Reverse Transcriptase Polymerase Chain Reaction (rRT-PCR) is the technique used in by the Center for Disease Control (CDC) to test for COVID-19. Real-time RT-PCR, or quantitative RT-PCR (RT-qPCR)*, is a specialized PCR technique that visualizes the amplification of the target sequence as it happens (in real-time) and allows you to measure the amount of starting target material in your reaction. You can read more about the basics of this technique, and watch a webinar here. For more about RT-PCR for COVID-19 testing, read this blog.
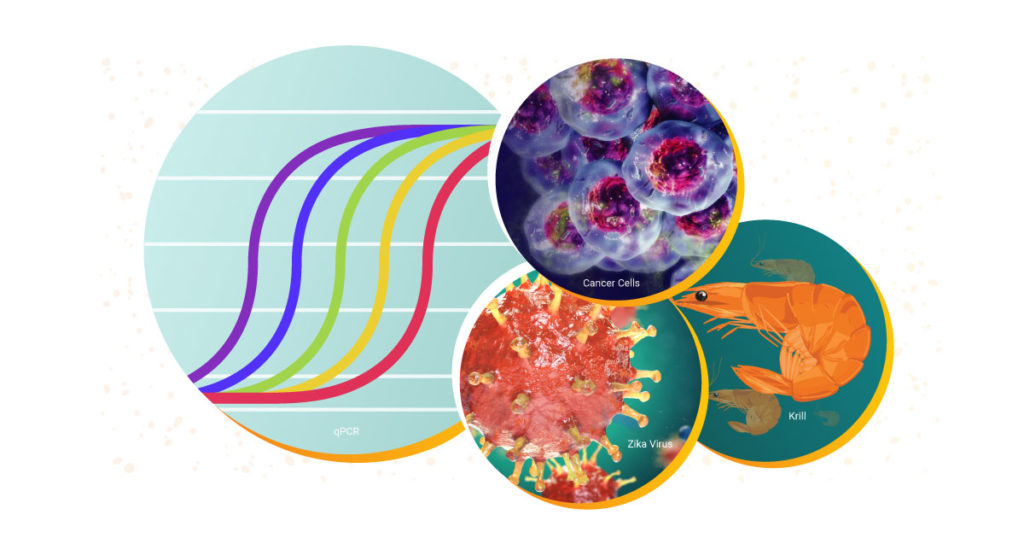
Both qPCR and RT-qPCR are powerful tools for scientists to have at their disposal. These fundamental techniques are used to study biological processes in a wide range of areas. Over the decades, Promega has supported researchers with RT-qPCR and qPCR reagents and systems to study everything from diseases like COVID-19 and cancer to viruses in elephants and the circadian rhythm of krill.
The Promega GoTaq® Probe 1-Step RT-qPCR System was added to a Centers for Disease Control and Prevention COVID-19 diagnostic protocol for emergency use during the SARS-CoV-2 pandemic. The GoTaq® Probe-based system for RT-qPCR offers scientists a ready-to-use mix of components that has been optimized for robust amplification. RT-qPCR assays are often used to measure the presence of viral RNA and to quantitate viral levels. For example, in this study of porcine epidemic diarrhea coronavirus (PEDV), the authors used viral RNA levels to demonstrate that after receptor binding, trypsin-dependent cleavage of the Spike (S) glycoprotein was necessary for cell entry and release of virions. To do this, they used the GoTaq® 1-Step RT-qPCR System to quantify the viral RNA released from infected cells inoculated in the presence or absence of trypsin (1).
Noronha et al. (2) and Venceslau et al. (3) measured transplacental transmission of Zikha virus using the GoTaq® 1-Step RT-qPCR System for viral RNA detection. Noronha et al. used RNA extracted from FFPE placental and necropsy brain tissue samples to demonstrate transmission of the Zika from mother to fetus (2). Venceslau et al showed the Zika virus can infect different regions of the placenta and umbilical cord by measuring viral RNA found in placental, umbilical cord and amniotic membrane samples (3).
The utility of RT-qPCR and qPCR extends well past the world of infectious disease, these techniques can also be used to measure gene expression levels. In this study, the expression of BCOR gene abnormalities in clear cell sarcomas of the kidney (CCSK) was measured using RT-qPCR. Clear cell sarcomas of the kidney are characterized by BCOR gene abnormalities. The authors used the GoTaq® 1-Step RT-qPCR System to measure the BCOR transcript expression of different BCOR gene abnormalities found in CCSK FFPE samples (4).
In this study, the upregulation of bone morphogenic protein 4 (BMP4) expression was measured with qPCR. BMP4 signaling is important in inhibiting neural differentiation and promoting the epidermal commitment of embryonic stem cells. The authors used the GoTaq® qPCR master mix to evaluate the effectiveness of additives to induce differentiation of human-induced pluripotent stem cells into corneal epithelial progenitor cells (5).
In a different application of qPCR to measure gene expression, it was used to shed greater light on the circadian rhythms of the Antarctic krill. Alberto Biscontin, one of the winners of the 2019 Promega real-time PCR grant, used GoTaq® qPCR Master Mix to validate expression of 10 clock control genes for his study. His qPCR data showed support for internal mechanisms that not only support daily living but also clarified the overwintering process of the krill. This study provided insight into the circadian transcriptome of these creatures who play a pivotal role in the Southern Ocean ecosystem, shedding light on the molecular mechanisms that help control and regulate their metabolic, physiological and behavioral rhythms (6).
All of these studies highlight the importance of qPCR and RT-qPCR to scientific research. They are “go-to” tools in scientist’s toolboxes, and can be applied in different ways to answer different questions. Foundational techniques such these help scientists and their research move forward, whether it is to battle a pandemic or understand the inner clock of krill, the answers they find help us better understand the world we live in.
* Due to differences within scientific literature, there are several acronyms describing PCR technologies utilizing reverse-transcriptase and offering real-time quantitation of DNA. The acronym implemented by the CDC, rRT-PCR, is equivalent to Promega RT—qPCR (Reverse Transcriptase quantitative Polymerase Chain Reaction).
References
- Wicht, O. et al .(2014) J. Virol. 88, 7952–61.
- Noronha, L.d. et al. (2016) Rio de Janeiro, 111, 287–93.
- Venceslau, E.M. et al. (2020) Front. Microbiol, 11, 112.
- Wong, M.K. et al. (2018) Histopathology. 72, 320–29.
- Kamarudin, T.A. et al. (2018) Stem Cells. 36, 337–48.
- Biscontin, A. et al. (2019) Sci. Rep. 9, 13894.
We’re committed to supporting scientists who are using molecular biology to make a difference. Learn more about our qPCR Grant program.
Related Posts
Kelly Grooms
Latest posts by Kelly Grooms (see all)
- The Battle of Shiloh’s Angel’s Glow: Fact, Civil War Legend or Modern Myth? - July 11, 2024
- Mind Control, Mutilation and Death. The Fungal Fate That Lurks in Waiting for Emerging Periodical Cicadas - June 13, 2024
- Measles and Immunosuppression—When Getting Well Means You Can Still Get Sick - May 13, 2024
