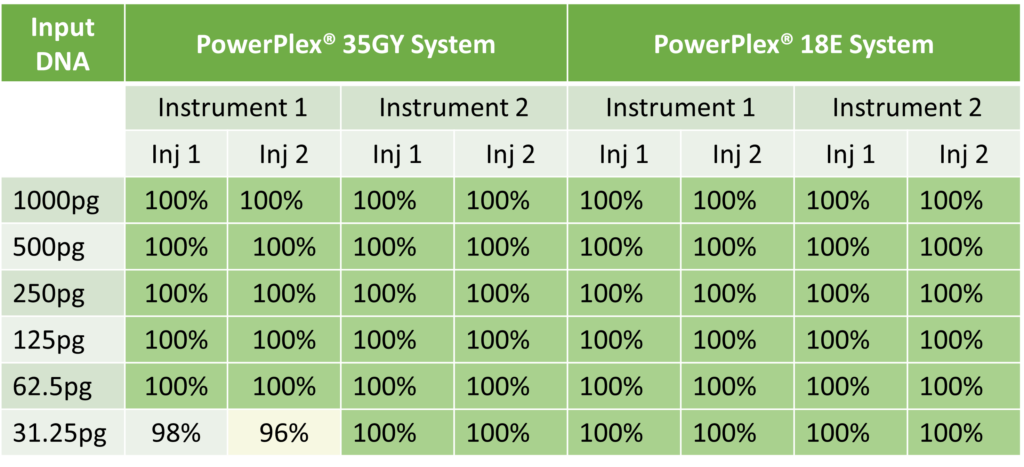
Almost three-quarters of the major crop plants across the globe depend on some kind of pollinator activity, and over one-third of the worldwide crop production is affected by bees, birds, bats, and other pollinators such as beetles, moths and butterflies (1). The economic impact of pollinators is tremendous: Between $235–577 billion dollars of global annual food production relies on the activity of pollinators (2). Nearly 200,000 species of animals act as pollinators, including some 20,000 species of bees (1). Some of the relationships between pollinators and their target plants are highly specific, like that between fig plants and the wasps that pollinate them. Female fig wasps pollinate the flowers of fig plants while laying their eggs in the flower. The hatched wasp larvae feed on some, but not all, of the seeds produced by fertilization. Most of the 700 fig plants known are each pollinated by only one or a few specific wasp species (3). These complex relationships are one reason pollinator diversity is critical.
Measuring the Success of Conservation Legislation

We are now beginning to recognize how critical pollinator diversity is to our own survival, and many governments, from the local level to the national level are enacting policies and legislation to help protect endangered or threatened pollinator species. However, ecosystems and biodiversity are complex subjects that make measuring and attributing meaningful progress on conservation difficult. Not only are there multiple variables in every instance, but determining the baseline starting point before the legislation is difficult. However, there are dramatic examples of success in saving species through legislative and regulatory action. The recovery of the bald eagle and other raptor populations in the United States after banning the use of DDT is one such example (4).
Continue reading “The Buzz on Biodiversity: Exploring Pollinator Diversity Through Mitochondrial DNA Analysis”