No protein is an island. Within a cell, protein-protein interactions (PPIs) are involved in highly regulated and specific pathways that control gene expression and cell signaling. The disruption of PPIs can lead to a variety of disease states, including cancer.
Two general approaches are commonly used to study PPIs. Real-time assays measure PPI activity in live cells using fluorescent or luminescent tags. A second approach includes methods that measure a specific PPI “after the fact”; popular examples include a reporter system, such as the classic yeast two-hybrid system.
Typically, the yeast two-hybrid system is used to screen a protein of interest against a library of potential binding partners. The system is based on the interaction of the yeast transcriptional activator GAL4 with a specific upstream activating sequence (UAS). The experimental method involves reconstitution of GAL4 activity from two separate constructs—the GAL4 DNA-binding domain is expressed as a fusion with protein “A”, while the transcriptional activation domain is fused to protein “B”. If the two fusion proteins interact, a functional GAL4 protein activates the UAS, driving expression of a reporter gene.
Previously, Kim et al. (1) reported a variation of the yeast two-hybrid system known as “specific protein association tool giving transcriptional readout with rapid kinetics” (SPARK). Their system included light gating—a mechanism based on a light-oxygen-voltage-sensing (LOV) domain containing a flavin chromophore that responds to blue light (450nm). Activation of the chromophore results in a covalent interaction that alters the conformation of the LOV domain. SPARK relies on both a PPI and light to activate transcription of the reporter gene (Figure 1A); thus, the activation event can be measured during a user-selected time window controlled by the delivery of blue light. SPARK introduces an additional variation on the two-hybrid system, as an alternative to reconstitution of GAL4. LOV domain activation by light and the interaction of proteins A and B allow tobacco etch virus protease (TEVp) to bind to its cleavage sequence (TEVcs), thereby liberating GAL4 and activating reporter gene expression (Figure 1B).
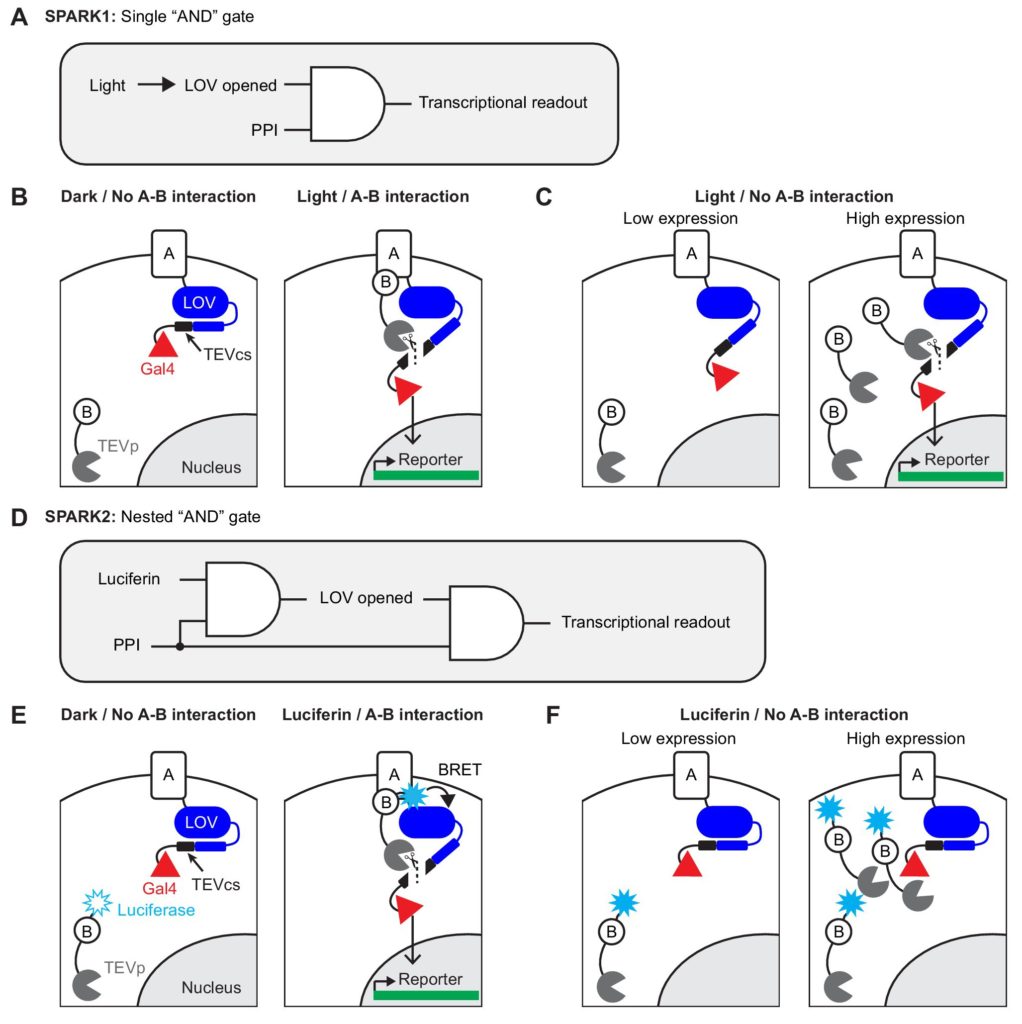
However, the original SPARK method suffered from two limitations. First, the system produced significant background signal when one of the fusion protein components was expressed at high levels—for example, during transient transfection (Figure 1C). Second, the delivery of blue light between intervals of darkness posed a practical challenge for automated systems, such as in high-throughput screening experiments.
If only there were a convenient source of blue light that could switched on when needed, within a cell…
Fortunately, there is. NanoLuc® luciferase is an engineered form of the luciferase gene found in the deep-sea shrimp Oplophorus gracillirostrus. It emits light at 460nm, close enough to the optimal wavelength required by the chromophore in the SPARK system.
In a recent publication (2), Kim et al. describe the development of a second-generation assay, SPARK2, that incorporates NanoLuc® luciferase to activate the LOV domain via bioluminescence resonance energy transfer (BRET).
In contrast to the original SPARK1, the authors describe SPARK2 as a “nested AND” logic gate (Figure 1D). Both furimazine (the substrate for NanoLuc® luciferase) and a PPI are required to induce a conformational change in the LOV domain which, in turn, leads to activation of the GAL4 UAS and consequent expression of the reporter gene (Figure 1E). SPARK2 overcomes the high background of the original SPARK method in the presence of high levels of protein B (Figure 1F), due to highly efficient and proximity-dependent activation of the LOV domain by light emitted from the NanoLuc® reaction. Additional experiments demonstrated that using furimazine as the “gating agent” in SPARK2 resulted in higher PPI specificity measurements as compared to light gating in SPARK1. This higher specificity was due to the nature of the intermolecular BRET interaction as part of the nested AND gate mechanism.
Further extending the utility of SPARK2, the authors demonstrated an orthogonal luciferase reporter system for high-throughput screening of G-protein coupled receptor (GPCR) agonists. This system employed firefly luciferase (FLuc) as the reporter gene driven by the GAL4 UAS, while the NanoLuc® BRET interaction mediated activation of the LOV domain as before. They did not observe cross-over luminescence between NanoLuc® and FLuc. They also provided a proof-of-concept demonstration that SPARK2 can be extended to study transcriptional readout of PPIs involved in surface contacts between cells.
In summary, SPARK2 is a highly sensitive and specific bioluminescent method to study PPIs within a cell, as well as those involved in intercellular interactions.
References
- Kim M.W. et al. (2017). Time-gated detection of protein-protein interactions with transcriptional readout. eLife 6:e30233. DOI: https://doi.org/10.7554/eLife.30233.
- Kim C.K., et al. (2019). Luciferase-LOV BRET enables versatile and specific transcriptional readout of cellular protein-protein interactions. eLife 2019;8:e43826. DOI: https://doi.org/10.7554/eLife.43826
Latest posts by Ken Doyle (see all)
- Will Artificial Intelligence (AI) Transform the Future of Life Science Research? - February 1, 2024
- RAF Inhibitors: Quantifying Drug-Target Occupancy at Active RAS-RAF Complexes in Live Cells - September 5, 2023
- Synthetic Biology: Minimal Cell, Maximal Opportunity - July 25, 2023
