This blog was written by guest author, Amy Landreman, PhD.
Drug repurposing, identifying new uses for approved or investigational drugs, is an attractive strategy when looking for new disease treatments. Because the compounds have already gone through some level of pre-clinical optimization and safety testing, this approach can reduce risk, reduce cost, and speed up the timeline for further drug development. An additional benefit of this approach is that it can result in new biological insights or a better understanding of disease mechanisms since these compounds usually already have some level of mechanistic characterization. Indeed, there are now a number of compound collections openly available specifically for the purpose of facilitating drug repurposing efforts. For example, the ReFRAME (Repurposing, Focused Rescue, and Accelerated Medchem) library is a collection of 12,000 compounds developed by Scripps Research Center and has been screened to identify novel candidate therapeutics for Cryptosporidium infection (1). The Broad Institute also offers a drug repurposing hub that contains an annotated collection of over 7,000 compounds.

Drug repurposing libraries, although often smaller than novel compound small molecule libraries, are designed for implementation into high-throughput screening workflows in order to efficiently triage compounds for the desired result. Effective compound screens require assays that can be scaled to 384 or 1536-well microplate formats and implemented in batch or continuous processing workflows. The firefly luciferase reaction has been leveraged to create many assays that are well-suited to these types of high-throughput screening approaches. In particular, the generation of “Glow” assays that have stable luminescent signals and homogenous assay design is a good fit. The signal stability allows for multi-plate processing and because the reagent is added directly to cells in culture, pre-processing steps are eliminated allowing for automated workflows. Assay reagents such as the CellTiter-Glo® Cell Viability Assay and the ADP-Glo™ Kinase Assay are commonly used in screening efforts including those done with repurposing libraries. In addition, there are several firefly luciferase reporter assay reagents such as Steady-Glo® and Bright-Glo™ Luciferase Assays that have been optimized for high-throughput detection of firefly luciferase activity making them well-suited to repurposing screens.
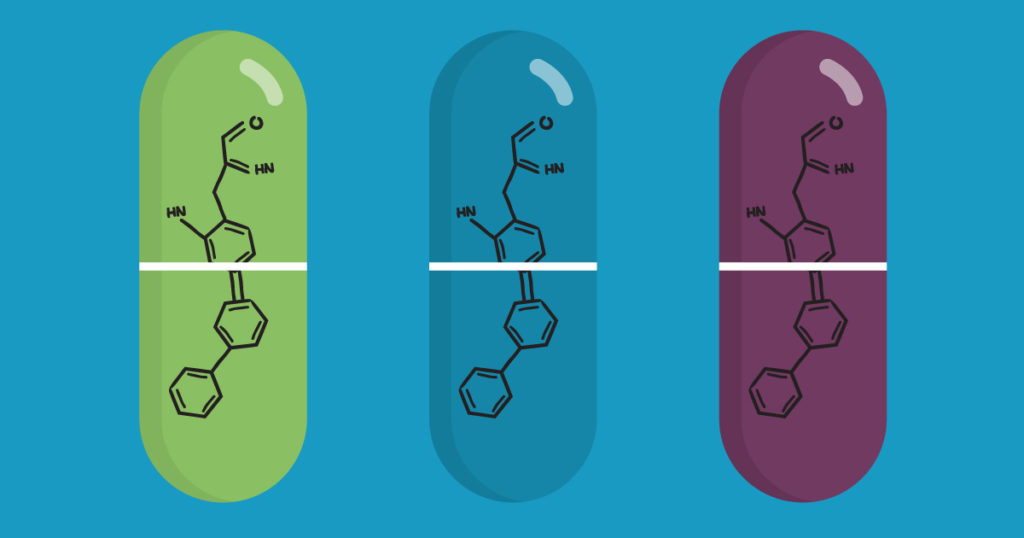

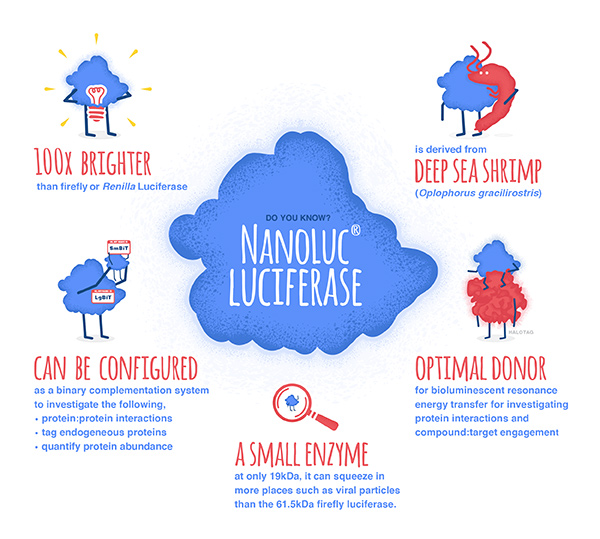

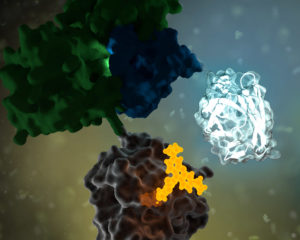 No protein is an island. Within a cell, protein-protein interactions (PPIs) are involved in highly regulated and specific pathways that control gene expression and cell signaling. The disruption of PPIs can lead to a variety of disease states, including cancer.
No protein is an island. Within a cell, protein-protein interactions (PPIs) are involved in highly regulated and specific pathways that control gene expression and cell signaling. The disruption of PPIs can lead to a variety of disease states, including cancer.