
Discovered in 1983 and initially dismissed as ‘cellular dust,’ exosomes have since emerged as pivotal players in biomedical research due to their roles in intercellular communication, potential as drug delivery vectors and as biomarkers for various diseases. These small extracellular vesicles, measuring 30–150nm, are crucial for transferring proteins, lipids, and nucleic acids — including microRNA (miRNA), mRNA, and non-coding RNA– between cells (1). miRNAs are particularly critical as they regulate gene expression and offer insights into the cellular mechanisms underlying diseases like cancer, enhancing the value of exosomes in cancer research.
Beyond exosomes importance in understanding intracellular communication and organ cross-talk, exosomes can also alter the functions of recipient cells based on their cargo. This capability makes them extremely valuable in providing insights into alterations in cellular communication, tumor microenvironments, metastasis and immune evasion.
Continue reading “What do Exosomes have to do with Cancer Research?”
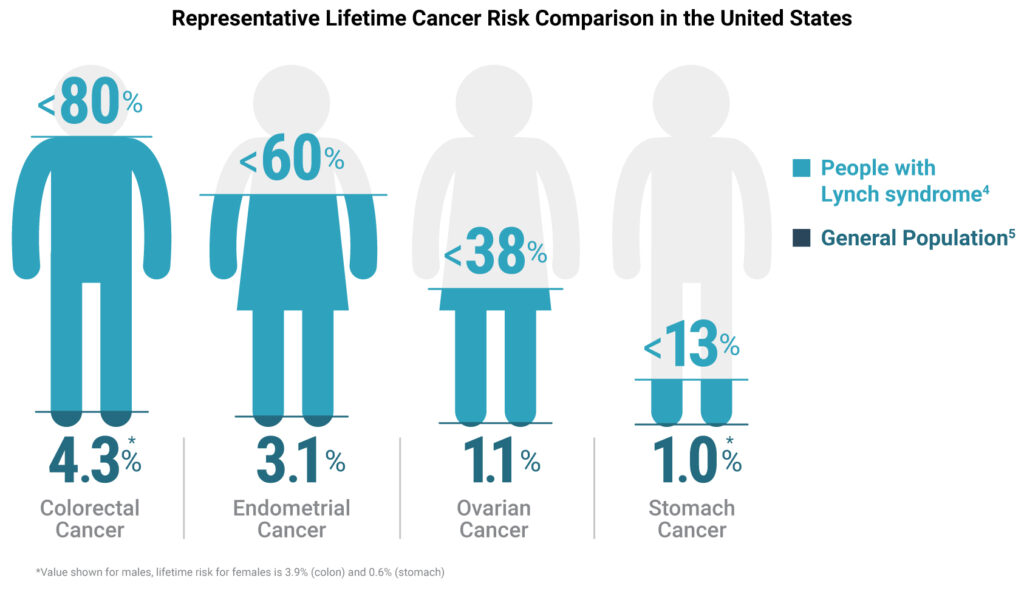
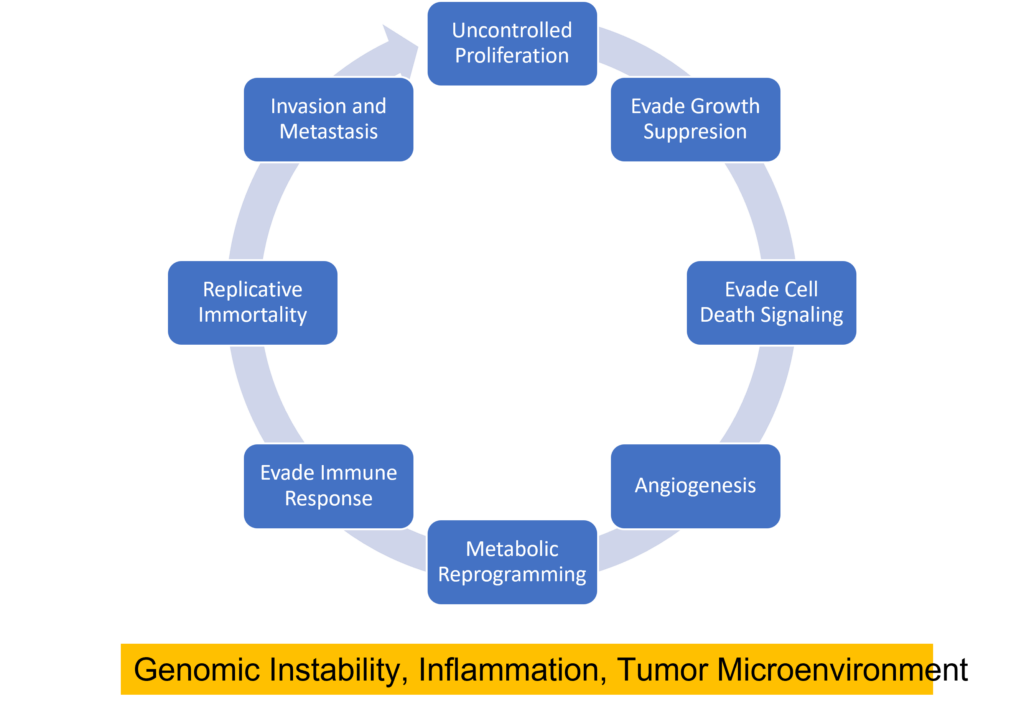


 When it comes to combating cancer does size matter? If every cell in the body has the propensity to become cancerous, it should naturally follow that larger animals that pack greater number of body cells and that those whose cells undergo greater number of cell divisions are more likely to develop cancer. By the same logic, organisms with longer lifespans must also have a greater chance of accumulating mutations leading to cancer. Surprisingly, the risk of developing cancer is only 5% in elephants and 18% in whales whereas it is as high as 30% in humans and rodents. The apparent lack of correlation between body mass, longevity and cancer- known as Peto’s paradox- has flummoxed scientists for several decades.1
When it comes to combating cancer does size matter? If every cell in the body has the propensity to become cancerous, it should naturally follow that larger animals that pack greater number of body cells and that those whose cells undergo greater number of cell divisions are more likely to develop cancer. By the same logic, organisms with longer lifespans must also have a greater chance of accumulating mutations leading to cancer. Surprisingly, the risk of developing cancer is only 5% in elephants and 18% in whales whereas it is as high as 30% in humans and rodents. The apparent lack of correlation between body mass, longevity and cancer- known as Peto’s paradox- has flummoxed scientists for several decades.1