This blog was written by guest contributor Tian Yang, Associate Product Manager, Promega, in collaboration with Kristin Huwiler, Manager, Small Molecule Drug Discovery, Promega.
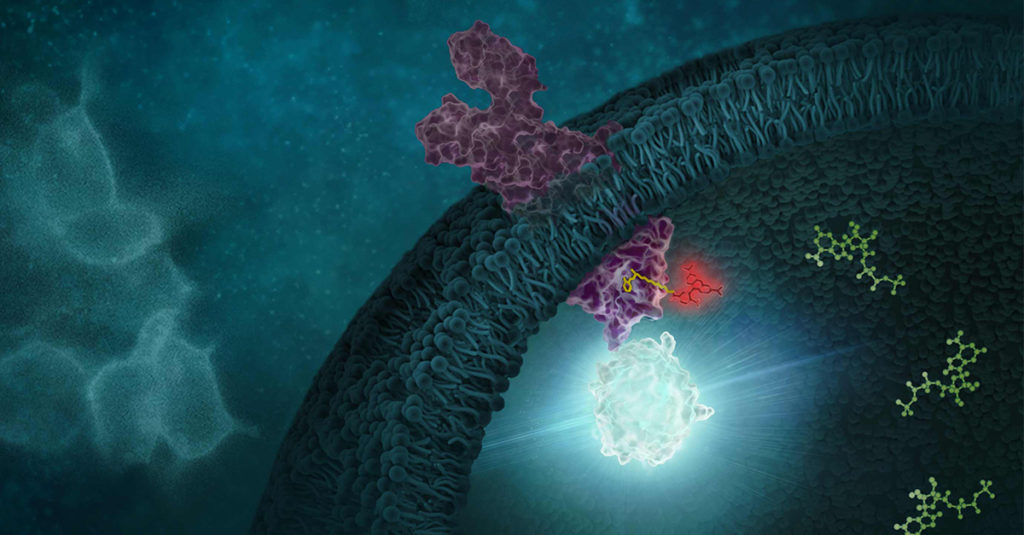
Determining the selectivity of a compound is critical during chemical probe or drug development. In the case of chemical probes, having a clearly defined mechanism of action and specific on-target activity are needed for a chemical probe to be useful in delineating the function of a biological target of interest in cells. Similarly, optimizing a drug candidate for on-target potency and reducing off-target interactions is important in the drug development process (1,2). A thorough understanding of the selectivity profile of a drug can facilitate drug repurposing, by enabling approved therapeutics to be applied to new indications (3). Interestingly, small molecule drugs do not necessarily require the same selectivity as a chemical probe, since some drugs may benefit from polypharmacology to achieve their desired clinical outcome.
Selectivity profiling panels based on biochemical methods have commonly been used to assess compound specificity for established target classes in drug discovery and chemical probe development. Biochemical assays are target-specific and often quantitative, enabling direct measurements of compound affinities for targets of interest and facilitate comparison of compound engagement to a panel of targets. As an example, several providers offer kinase selectivity profiling services using different assay formats and kinase panels comprised of 100 to 400 kinases (4). However, just as biochemical target engagement does not always translate to cellular activity, selectivity profiles based on biochemical platforms may not reflect compound selectivity in live cells (5).
Continue reading “Cellular Selectivity Profiling: Unveiling Novel Interactions and More Accurate Compound Specificity”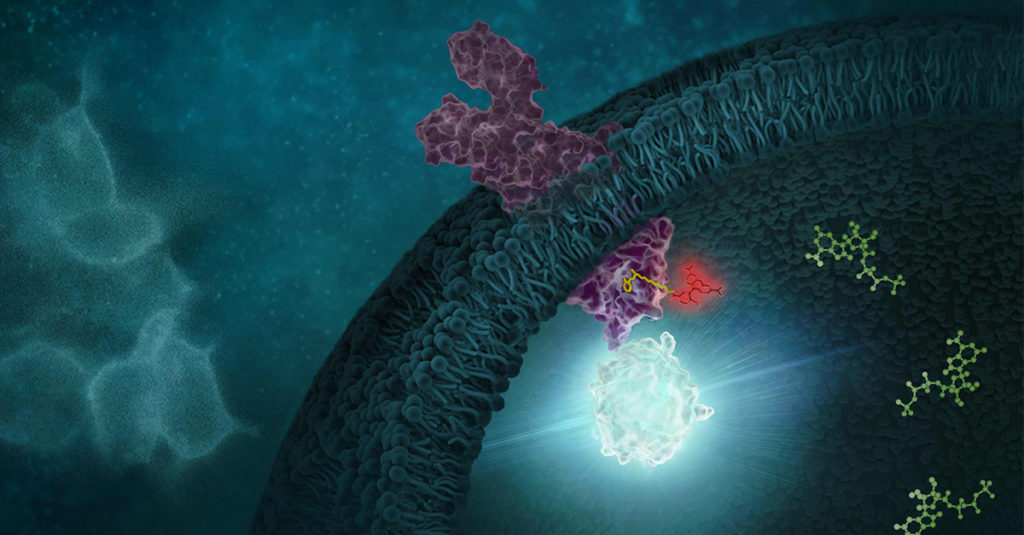

 Designing and implementing a new assay can be a challenging process with many unexpected troubleshooting steps. We wanted to know what major snags a scientist new to the
Designing and implementing a new assay can be a challenging process with many unexpected troubleshooting steps. We wanted to know what major snags a scientist new to the 