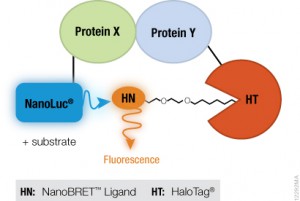
Cells, commonly considered the smallest unit of life, provide structure and function for all living things (3).
Because cells contain the fundamental molecules of life, in some situations such as yeast, a single cell can be considered the complete organism. In other situations, for more complex multicellular organisms, a multitude of cells can mature and acquire different, specialized functions (3).
Cells developing specificity are undergoing differentiation, a process where a cell’s genes are either turned “on” or “off” resultant in a more specific cell type. As these differentiated cells start to exhibit their identity, they organize themselves into the tissues, organs, and organ systems integral to the functioning of a multicellular, developing organism. This process in which order and form is created within a developing organism is referred to as morphogenesis (5).
Continue reading “Cell Tracking Using HaloTag: Why are Scientists Chasing Cells?”
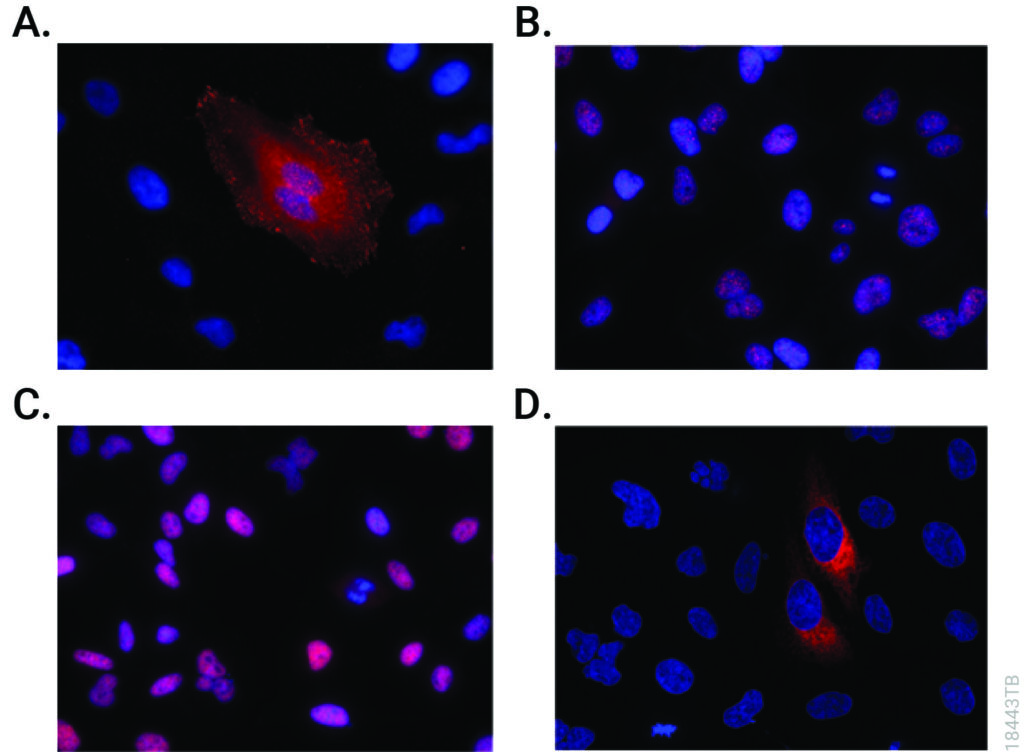
 Designing and implementing a new assay can be a challenging process with many unexpected troubleshooting steps. We wanted to know what major snags a scientist new to the
Designing and implementing a new assay can be a challenging process with many unexpected troubleshooting steps. We wanted to know what major snags a scientist new to the  Antibodies labeled with small molecules such as fluorophore, biotin or drugs play a critical role in various areas of biological research,drug discovery and diagnostics. There are several limitations to current methods for labeling antibodies including the need for purified antibodies at high concentrations and multiple buffer exchange steps.
Antibodies labeled with small molecules such as fluorophore, biotin or drugs play a critical role in various areas of biological research,drug discovery and diagnostics. There are several limitations to current methods for labeling antibodies including the need for purified antibodies at high concentrations and multiple buffer exchange steps.
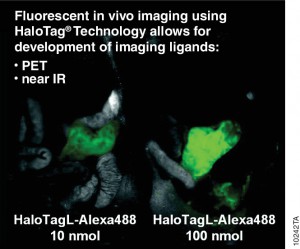 Antibodies labelled with radioisotopes or the sequential administrationof an antibody and a radioactive secondary agent facilitate the in vivo detection and/or characterisation of cancers by positron emission tomography (PET) or by single-photon emission computed tomography (SPECT) imaging.
Antibodies labelled with radioisotopes or the sequential administrationof an antibody and a radioactive secondary agent facilitate the in vivo detection and/or characterisation of cancers by positron emission tomography (PET) or by single-photon emission computed tomography (SPECT) imaging.