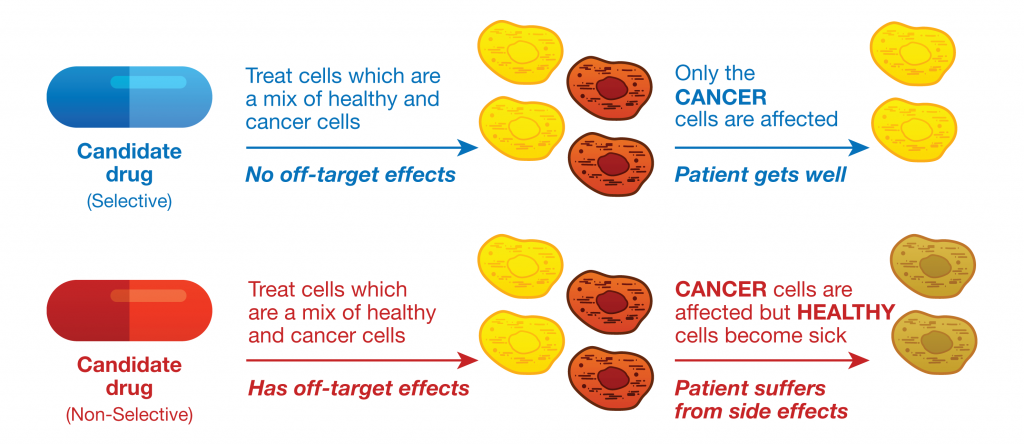
The understudied kinome represents a major challenge as well as an exciting opportunity in drug discovery. A team of researchers lead by Nathanael Gray at the Dana Farber Cancer Institute was able to partially elucidate the function of an understudied kinase, Doublecortin-like kinase 1 (DCLK1), in pancreatic ductal adenocarcinoma cells (PDAC). The characterization of DCLK1 in PDAC was realized by developing a highly specific chemical probe (1). Promega NanoBRET™ Target Engagement (TE) technology enabled intracellular characterization of this chemical probe.
The Dark Kinome
Comprised of over 500 proteins, the human kinome is among the broadest class of enzymes in humans and is rife with targets for small molecule therapeutics. Indeed, to date, over 50 small molecule kinase inhibitors have achieved FDA approval for use in treating cancer and inflammatory diseases, with nearly 200 kinase inhibitors in various stages of clinical evaluation (2). Moreover, broad genomic screening efforts have implicated the involvement of a large fraction of kinases in human pathologies (3). Despite such advancements, our knowledge of the kinome is limited to only a fraction of its family members (3,4). For example, currently less than 20% of human kinases are being targeted with drugs in clinical trials. Moreover, only a subset of kinases historically has garnered substantial citations in academic research journals (4). As a result, a large proportion of the human kinome lacks functional annotation; as such, these understudied or “dark” kinases remain elusive to therapeutic intervention (4).
Continue reading “Illuminating the Function of a Dark Kinase (DCLK1) with a Selective Chemical Probe”