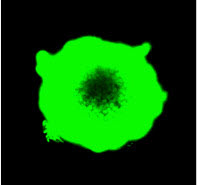
In 3D cell culture models, cells are grown under conditions that allow the formation of multicellular spheroids or microtissues. Instead of growing in a monolayer on a plate surface, cells in 3D culture grow within a support matrix that allows them to interact with each other, forming cell:cell connections and creating an environment that mimics the situation in the body more closely than traditional 2D systems. Although 3D cultures are designed to offer a more physiologically accurate environment, the added complexity of that environment can also present challenges to experimental design when performing cell-based assays. For example, it can be a challenge for assay reagents to penetrate to the center of larger microtissues and for lytic assays to disrupt all cells within the 3D system.
Earlier this week Terry Riss, a Senior Product Specialist at Promega, presented a Webinar on the challenges of performing cell-based assays on microtissues in 3D cell culture. During the Webinar, Terry gave an overview of the different methods available for 3D cell culture, providing a description of the advantages of each. He then discussed considerations for designing and optimizing cell-based assays for use in 3D culture systems, providing several recommendations to keep in mind when performing cell viability assays on larger microtissue samples.
Continue reading “3D Cell Culture Models: Challenges for Cell-Based Assays”