Targeting a single protein and making it disappear from the cell is quite the magic trick, and there are various molecular tools available for this task. You can use RNA interference, which prevents a protein from being made, inhibitors that bind the protein, rendering it unavailable for use or even gene editing tools like CRISPR that can remove the encoding gene from the genome. Also, you can target an existing protein for destruction, using the cell’s own garbage disposal system to degrade the protein, and you can even expand the concept of targeted degradation to other kinds of macromolecules, such as RNA.
Why is this important? There are many cellular molecules that play a role in diseases like cancer, but researchers are unable to find compounds that might block their activity. These molecules, like transcription factors such as MYC, are called “undruggable” because they have no binding site for small molecules to bind and inhibit function. However, if you could specifically target the troublesome molecule and essentially delete it from the cell, you could overcome the “undruggable” problem.
PROTACs Bring Destruction to Targeted Proteins
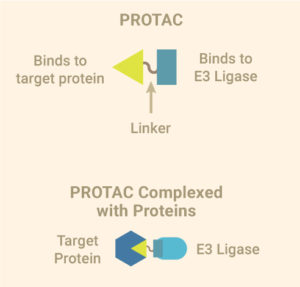
PROTAC or the proteolysis-targeting chimera is the first kind of tool developed for targeting specific proteins for degradation using the Ubiquitin Proteasome System (UPS). A PROTAC consists of three elements: A ligand that binds to a ubiquitin E3 ligase, a second ligand that binds the protein of interest and a linker that brings the two ligands together. The E3 ligase is part of a protein complex that tags proteins with ubiquitin, a molecular signal to clear away damaged proteins in the cell. By collecting the protein you are interested in and hitching it to the E3 ligase, the bifunctional PROTAC introduces the target protein to the protein complex that adds ubiquitin, which forces the target protein down the path to destruction. Ubiquitinated proteins are directed to the proteasome where they are broken down into their component molecules, ready to be recycled by the cell.
PROTACs opened the door to targeting proteins that couldn’t be reached with traditional drugs like small-molecule inhibitors or antibodies. In 2019, the first PROTAC-based drug candidate, ARV-110, entered clinical trials for prostate cancer, and ARV-471 is an estrogen receptor-targeting PROTAC for breast cancer. In March 2025, Phase III clinical trial data were positive.
PHOTACs: Making PROTACs Responsive to Light
PROTACs are bifunctional molecules. Here a paper describes a trifunctional molecule based on PROTACs that contains a “photoswitch”. By adding the photoswitch to the ligand for E3 ligase and the ligand for the protein target, Reynders et al. generated what they called photochemically targeted chimeras (PHOTACs) that are not active until exposed to light at 380-440nm. They tested their newly developed PHOTACs with bromodomain-containing proteins (BRD2-4) and FK506 binding protein 12 (FKPB12), proteins that have been successfully degraded by existing PROTACs. They observed degradation when the cells were exposed to light while those that remained in the dark maintained their protein levels. Because photoswitches are reversible, the cells dosed with the BRD PHOTAC were exposed first to the activating light wavelength and then the deactivating wavelength. The amount of BRD2 did decrease initially but recovered more quickly than cells that were exposed to the activating light and left in the dark.
Being able to activate the degradation of a particular protein at a particular time has advantages, including minimizing off-target effects (i.e., altering the degradation of a protein other than the protein of interest) and fine tuning control of when degradation occurs. PHOTAC reversibility means that you can also control how long the protein is targeted for degradation. The technique is new, but the authors expand on its possible applications.
Molecular Glues: Aiming at Small Intracellular Targets
Molecular Glues (sometimes called small-molecule PROTACs) use small molecules to drive protein:protein interactions. Specifically, molecular glues use the UPS system to drive protein degradation but do so by using small molecules to recruit endogenous substrate receptors of E3, like Cereblon (CRBN). These molecules are typically used to target intracellular proteins such as transcription factors or other difficult targets. Examples of molecular glues are iMiDs which are immunomodulatory drugs including thalidomide and other imides and are being used in the clinic to treat specific cancers, such as multiple myeloma.
LYTACs: A Different Path to Degradation
Because PROTACs act on proteins with domains exposed to the cytosol, there are membrane proteins and secreted proteins that elude this strategy. But what if you could target proteins to the lysosome, rather than the proteasome? That particular degradation method was described in an article as a way to target some of those proteins not served by the PROTACs. By exploiting a known lysosome targeting receptor [cation-independent mannose-6-phosphate receptor (CI-M6PR)] that directs proteins to the lysosome and an antibody specific to the protein, Banik et al. describe how they generated lysozyme targeting chimeras (LYTACs). To test their concept, they used a polypeptide with multiple ligands for CI-M6PR attached to biotin and examined if NeurAvidin was taken up by the cells. Avidin and biotin have a high affinity for each other. In the presence of the biotin LYTAC, the NeurAvidin was colocalized to endosomes and lysosomes.
Could they use an antibody to target a protein to the lysosome? After showing that they could conjugate glycoprotein ligands to an antibody, the LYTAC was incubated with a fluorescently labeled mouse IgG and the fluorescence was 40-fold higher in the lysosome relative to controls. This demonstrated an extracellular protein could be sent to the lysosome using the engineered LYTAC. If the protein was bound to its antigen, would the LYTAC still work? When using a fluorescent reporter mCherry bound to mouse anti-mCherry antibody or a protein implicated in neurodegenerative disease, apoliporotein E4 (ApoE4), bound to mouse anti-ApoE4 antibody with the LYTAC, there was at least an order of magnitude increase in lysosomal uptake of the protein compared to untreated cells.
Learn more about targeted protein degradation at the Promega.com website.
Would LYTACs also work for membrane proteins? Like with PROTACs, the membrane protein and CI-M6PR would have to bind the LYTAC simultaneously. To target epidermal growth factor receptor (EGFR), which plays a role in cancer proliferation, cetuximab, an EGFR-blocking antibody, was conjugated to a glycopeptide with multiple CI-M6PR ligands. When cells were treated with the EGFR LYTAC for 24 hours, the amount of EGFR was substantially decreased compared to control treatment, indicating the targeted protein had been degraded. When tested with other membrane proteins like programmed death-ligand 1 (PD-L1), a receptor overexpressed on cancer cells, the amount of PD-L1 on the cell surface decreased in two different cell lines: 33% decrease in a cell line that expressed CI-M6PR at low levels and 50% decrease in a Hodgkin’s lymphoma cell line with higher levels of CI-M6PR. Taking all the data together, the proof-of-concept by Banik et al. shows that LYTACs are an approach to target extracellular proteins for degradation in the lysosome.
AUTAC/ATTEC and Other Chaperone Degraders: Using the Autophagy-Lysosomal Pathway
These degraders recruit the autophagosome or chaperone-mediated autophagy (CMA), and the targets of these degraders are aggregated proteins, organelles like mitochondria, or possibly even large cytosolic disease proteins (such as the aggregates observed in many neurogenerative diseases). One example in research is application of the autophagosome-tethering compound (ATTEC) to target the accumulation of mHTT in cells.
Nucleic Acid Degrader (RIBOTAC): Going after Disease-Causing RNA Molecules
RNases or RNA-binding nucleases are recruited to target RNA, leading to cleavage of the RNA. These degraders are used to target disease-related RNAs, oncogenic long-codeing RNAs or difficult-to-inhibit mRNAs. One example in research is a RIBOTAC that degrades miRNA 21 by recuring RNAse L.
Conclusion
Targeting disease-causing molecules for degradation is a strategy to circumvent the difficult-to-drug targets that have known roles in diseases like cancer. Targeted degradation of these molecules is one approach researchers are taking to mitigate the effect a particular protein or other macromolecule has in a cellular pathway. Already this strategy has yielded therapeutics that are showing promise in treating cancers and other diseases.
Originally published in 2019. Updated June 30, 2025
Related Posts
Updates 3/10/21
Latest posts by Promega (see all)
- Accurate and On-Time: A Look Inside Promega Logistics - June 24, 2025
- Augmenting Human Capabilities with AI Tools - June 17, 2025
- Lead with Empathy: Supporting Caregivers in the Workplace - June 3, 2025
