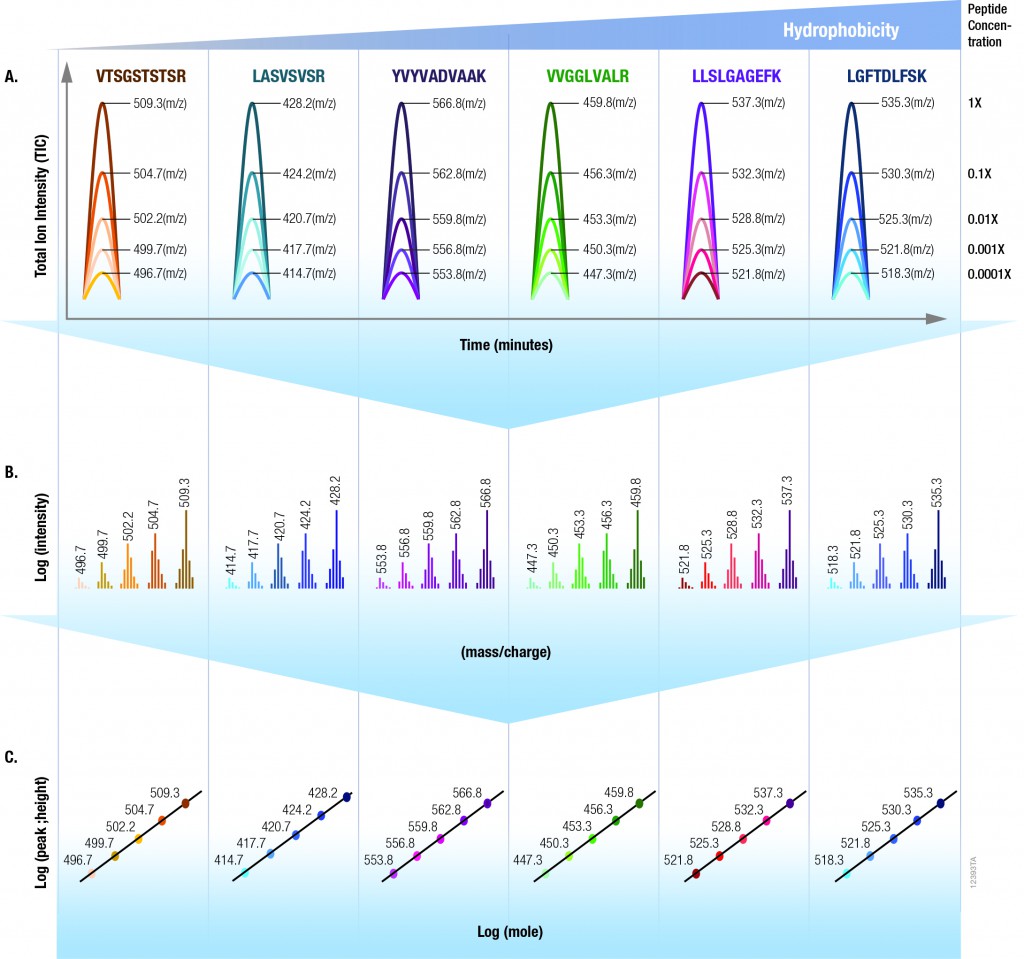
One goal of drug discovery and research programs is to reduce false hits as early as possible in the process. Follow-up on false hits is costly in terms of time and resources, and the longer the false hits remain in the drug development pipeline, the more costly they are. So methods that can easily reduce the number of false hits during compound screening early in the discovery process are particularly sought after.
Reporter assays have proven to be invaluable tools for elucidating the mechanisms of action of small molecules or other agents on signaling pathways within cells, and the luciferase reporter assay has become a standard research tool in the biological research laboratory.
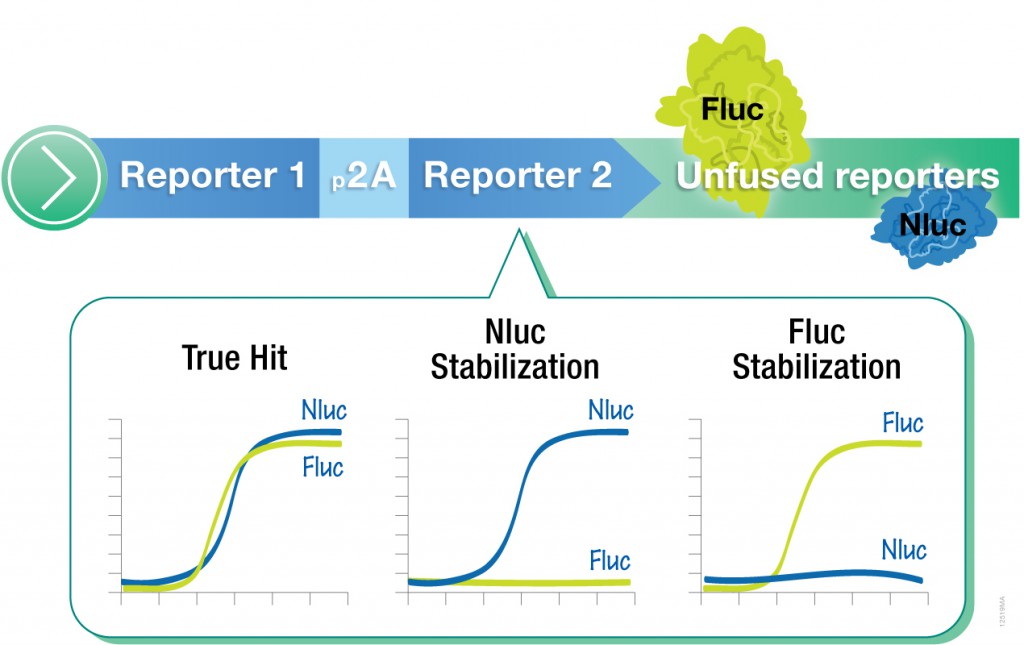
However, one caveat of using standard luciferase-based reporter assays for larger-scale compound screening efforts is the frequency of false hits that result from direct interaction of compounds with the luciferase reporter. This issue can be mitigated with a “coincidence reporter” system where two independent reporter proteins are produced from a single transcript. In this type of assay, a bicistronic transcript is stoichiometrically translated into two nonhomologous reporters by means of a 2A “ribosomal skipping” sequence. Since it is unlikely that compounds will interact with two distinct types of reporter, “coincident” responses will indicate on-target activity. Such a coincident reporter system provides an important control against costly false hits early in drug discovery research programs.
A paper published online in ACS Chem Biol in February describes the first successful application of the firefly/NanoLuc luciferase coincidence reporter system to identify new pathways that up-regulate PARK2 expression.


 Timing is everything! I learned that the hard way just two weeks ago when I took my son to scout camp and thought I would try to capture the traditional American flag ceremony for posterity. I set up my camera for a panoramic shot and scanned the crowd. Feeling very pleased with myself, I got home that evening ready to show my family the great camera skills I had honed over the Summer months. To my horror, I noticed that half of the scout troop was saluting the flag while the other half were standing to attention! I had got the timing horribly wrong (although the picture is still fun to look at in a strange sort of way).
Timing is everything! I learned that the hard way just two weeks ago when I took my son to scout camp and thought I would try to capture the traditional American flag ceremony for posterity. I set up my camera for a panoramic shot and scanned the crowd. Feeling very pleased with myself, I got home that evening ready to show my family the great camera skills I had honed over the Summer months. To my horror, I noticed that half of the scout troop was saluting the flag while the other half were standing to attention! I had got the timing horribly wrong (although the picture is still fun to look at in a strange sort of way).