On July 3rd and 4th, 2021, we celebrate World Firefly Day, and 2021, marks 30 years of luciferase products firefly luciferase vectors and Luciferase Assay System. These tools are key in advancing bioluminescent technology. To celebrate this day, we want to highlight some innovations that have been made possible with these tools.
Continue reading “World Firefly Day: Shining More Light on Glo-ing Innovations”luciferase reporter
Firefly Luciferase Sheds Light on Development of New Malaria Treatments

Despite significant advancements in antimalarial drugs and widespread efforts to prevent transmission over the past decade, deaths from malaria remain high, particularly in younger children. New drugs with novel modes of action are urgently needed to continue reducing mortality and address drug resistance in the malaria parasite, Plasmodium falciparum. While tens of thousands of compounds have been identified as potential candidates through massive screening efforts, scalable methods for identifying the most effective compounds are needed.
The goal is to find a drug that is potent during all stages in the life cycle of P. falciparum and kills the parasite quickly. Focusing on assessing whether a compound can rapidly eliminate initial parasite burden, Paul Horrocks, PhD, and his colleagues developed a validated bioluminescence-based assay that rapidly determines the initial rate of kill for discovery antimalarials. One key to developing their assay was figuring out how to monitor when the parasite dies after introducing the drug. While measuring DNA content can be used to monitor parasite burden, it is too stable to use for a relevant time course assay.
Enter firefly luciferase, a dynamic reporter tool to investigate drug action. By creating transgenic P. falciparum that express the luc reporter gene, the researchers could monitor drug action over time. When the parasite is killed, it stops making the luciferase reporter. Since there is no new production of luciferase, levels fall quickly after the parasite dies, and a luciferase assay can determine how fast each drug killed the parasite.
Continue reading “Firefly Luciferase Sheds Light on Development of New Malaria Treatments”Bioluminescence and Biotechnology: Shining Nature’s Cool Light on Biology
Imagine you’re taking a refreshing night swim in the warm blue waters of Vieques in Puerto Rico. You splash into the surf and head out to some of the deeper waters of the bay, when what to your wondering eyes should appear, but blue streaks of light in water that once was clear. Do you need to get your eyes checked? Are you hallucinating? No! You’ve just happened upon a cluster of dinoflagellates, harmless bioluminescent microorganisms called plankton, that emit their glow when disturbed by movement. These dinoflagellates are known to inhabit waters throughout the world but are generally not present in large enough numbers to be noticed. There are only five ecosystems in the world where these special bioluminescent bays can be seen, and three of them are in Puerto Rico.

But you don’t have to travel to Puerto Rico or swim with plankton to see bioluminescence. There are bioluminescent organisms all over the world in many unexpected places. There are bioluminescent mushrooms, bioluminescent sea creatures—both large and small (squid, jellyfish, and shrimp, in addition to the dinoflagellates)—and bioluminescent insects, to name a few. Bioluminescence is simply the ability of living things to produce light.
Continue reading “Bioluminescence and Biotechnology: Shining Nature’s Cool Light on Biology”I Have My Luciferase Vector, Now What?
Choosing and Optimizing Transfection Methods
Here in Technical Services we often talk with researchers at the beginning of their project about how to carefully design and get started with their experiments. It is exciting when you have selected the Luciferase Reporter Vector(s) that will best suit your needs; you are going to make luminescent cells! But, how do you pick the best way to get the vector into your cells to express the reporter? What transfection reagent/method will work best for your cell type and experiment? Do you want to do transient (short-term) transfections, or are you going to establish a stable cell line?
Continue reading “I Have My Luciferase Vector, Now What?”NanoBiT™ Assay: Transformational Technology for Studying Protein Interactions Named a Top 10 Innovation of 2015

For three out of the last four years, we have been honored to have one of our key technologies named a Top 10 Innovation by The Scientist. This year the innovative NanoBiT™ Assay (NanoLuc® Binary Technology) received the recognition. NanoBiT™ is a structural complementation reporter based on NanoLuc® Luciferase, a small, bright luciferase derived from the deep sea shrimp Oplophorus gracilirostris.
Using plasmids that encode the NanoBiT complementation reporter, you can make fusion proteins to “report” on protein interactions that you are studying. One of the target proteins is fused to the 18kDa subunit; the other to the 11 amino acid subunit. The NanoBiT™ subunits are stable, exhibiting low self-affinity, but produce an ultra-bright signal upon association. So, if your target proteins interact, the two subunits are brought close enough to each other to associate and produce a luminescent signal. The strong signal and low background associated with a luminescent system, and the small size of the complementation reporter, all help the NanoBiT™ assay overcome the limitations associated with traditional methods for studying protein interactions.
The small size reduces the chances of steric interference with protein interactions. The ultra bright signal, means that even interactions among proteins present in very low amounts can be detected and quantified–without over-expressing large quantities of non-native fusion proteins and potentially disrupting the normal cellular environment. And the NanoBiT™ assay can be performed in real time, in live cells.
The NanoBiT™ assay is already being deployed in laboratories to help advance understanding of fundamental cell biology. You can see how one researcher is already taking full advantage of this innovative technology in the video embedded below:
Visit the Promega web site to see more examples more examples how the NanoBiT™ assay can break through the traditional limitations for studying protein interactions in cells.
You can read the Top 10 article in The Scientist here.
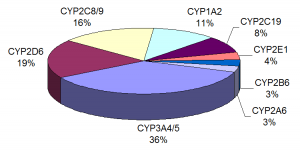
Screening for Drug-Drug Interactions with PXR and CYP450 3A4 Activation
The pregnane X receptor (PXR) is a nuclear receptor known to regulate expression of cytochrome P450 (CYP450) drug-metabolizing enzymes (1). PXR has even been designated the “master xenosensor” due to its ability to upregulate cellular levels of a variety of drug-metabolizing enzymes in response to drugs and foreign chemicals. Elevated levels of CYP450 enzymes can elicit alterations in the pharmacokinetics of co-administered drugs, which can result in adverse drug-drug interactions (DDI) or diminished bioavailability. By assessing PXR activation and CYP450 enzyme induction early in the drug development process, many companies hope to reduce late-stage clinical failures and minimize the high costs associated with bringing a new drug to market.
A paper by Shukla et al. (2) examined over 2,800 clinically used drugs for their ability to activate human PXR (hPXR) and rat PXR (rPXR), induce human cytochrome P450 3A4 enzyme (CYP3A4) at the cellular level, and bind hPXR at the protein level. Several studies have identified PXR as playing a key role in regulating the expression of CYP3A4, an enzyme involved in the metabolism of more than 50% of all drugs prescribed in humans. Since PXR activation and CYP3A4 induction have an impact on drug metabolism and pharmacokinetics, the authors wanted to obtain data that would be valuable in understanding structure-activity relationships (SARs), the connection between chemical structure and biological activity, when prioritizing new molecular entities (NMEs) for further in vitro and in vivo studies.
Continue reading “Screening for Drug-Drug Interactions with PXR and CYP450 3A4 Activation”