This blog is written by guest blogger, Heather Tomlinson, former Director of Clinical Diagnostics at Promega.
Finding safe and effective treatments for human diseases takes time. Medication and diagnostic tests can take decades to discover, develop and prove safe and effective. In the United States, the FDA stands as the gold-standard gatekeeper to ensure that treatments and tests are reliable and safe. The time we wait in review and clearance means less risk of ineffective or unsafe treatments.
And yet, in a pandemic, we are behind before we even start the race to develop diagnostic tests, so critical for understanding how an infectious disease is spreading. That is when processes like the FDA’s fast track of Emergency Use Authorization (EUA) are critical. Such authorization allows scientists and clinicians to be nimble and provide the best possible test protocol as quickly as possible, with the understanding that these protocols will continue to be evaluated and improved as new information becomes available. The EUA focuses resources and accelerates reviews that keep science at the fore and gets us our best chance at staying safe and healing.

For scientists working around the clock, the FDA’s EUA process is ready to review and respond. Getting an EUA gives clinical labs a very specific and tested resource to guide them to the tools and tests to use in a crisis.
Typically the Centers for Disease Control (CDC) will develop the first test or protocol that receives FDA EUA in response to a crisis like a pandemic. For COVID-19 the CDC 2019-Novel Coronavirus Real-Time RT-PCR Diagnostic Panel received FDA EUA clearance in early February. This is the test protocol used by the public health labs that work with the CDC and test manufacturers around the world.
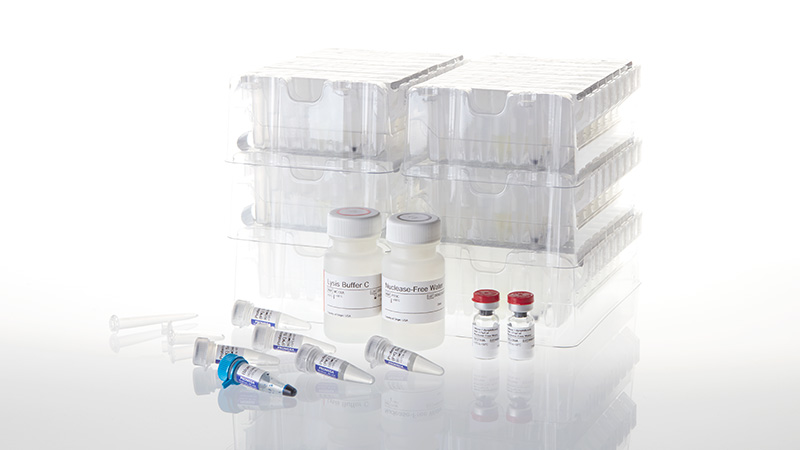
Throughout a crisis such as the current pandemic, scientists continually work to improve the testing protocols and add options to the EUA protocols. This enables more flexibility in the test protocols. Promega is fortunate to play a part of the CDC EUA equation for diagnostic testing. Our GoTaq® Probe 1-Step PRT-qPCR System is one of a few approved options for master mixes in the CDC qPCR diagnostic test, and now our medium-throughput Maxwell 48 Instrument and Maxwell Viral Total Nucleic Acid Purification Kit were added to the CDC protocol as an option for the RNA isolation step as well. These additions to the CDC EUA means that laboratories have more resources at their disposal for the diagnostic testing which is so critical to effective pandemic response.
The Emergency Use Authorization provides the FDA guidance to strengthen our nation’s public health during emergencies, such as the current COVID-19 pandemic. The EUA allows continual improvement of an authorized protocol through the collaborative efforts scientists in all academia, government and industry to identify and qualify the most reliable technologies and systems, giving labs more flexibility as new products are added as options.

Dr. Tomlinson was the Director for the Global Clinical Diagnostics Strategic Business Unit at Promega Corporation bringing over 15 years of experience in clinical diagnostic test development. She was responsible for leading the team that drives strategy in the clinical market for Promega. Her background was in infectious disease diagnostic testing, with a focus on HIV drug resistance and evolution. Her last work focused on oncology companion diagnostic test development. Heather was an accomplished international presenter, delivering conference presentations in the United States, Europe, Asia, and Africa. Heather passed away in 2023.