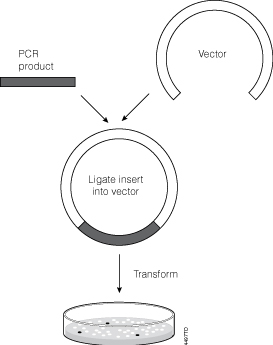
 Let’s face it, most lab techs and purchasing agents aren’t all that happy when you send them an Instagram picture of your latest lunchroom-napkin cloning strategy as your order form for your next big cloning experiment. So we have created the CloneWeaver® Workflow Builder. You can transfer your brilliance easily from that lunchroom napkin to an orderly email or print out of every vector, enzyme, purification kit, and transfection reagent your next big molecular cloning experiment requires. You can even save your one-of-a-kind “cloning kit” for future endeavors.
Let’s face it, most lab techs and purchasing agents aren’t all that happy when you send them an Instagram picture of your latest lunchroom-napkin cloning strategy as your order form for your next big cloning experiment. So we have created the CloneWeaver® Workflow Builder. You can transfer your brilliance easily from that lunchroom napkin to an orderly email or print out of every vector, enzyme, purification kit, and transfection reagent your next big molecular cloning experiment requires. You can even save your one-of-a-kind “cloning kit” for future endeavors.
The CloneWeaver® tool will walk you through every step of the molecular cloning process from selecting a vector to finding a transfection reagent for mammalian cells. So if you are starting a new project, we are with you every step of the way. We will help you find restriction enzymes and even remind you about markers and biochemicals that you may want to have on hand for your experiment. Within the tool we have links to additional resources like our RE Tool and catalog pages if you need more help.
 Already have a favorite vector and a freezer full of restriction enzymes? No problem, skip those steps and move on to getting the perfectly sized nucleic acid markers or the particular polymerase your experiment requires.
Already have a favorite vector and a freezer full of restriction enzymes? No problem, skip those steps and move on to getting the perfectly sized nucleic acid markers or the particular polymerase your experiment requires.
Are you teaching a molecular genetics course? CloneWeaver® workflow builder is perfect for creating the list of laboratory reagents you are going to need for your students—and you will have this same list as a starting point for other lab experiments or classes later on because you can save the lists that you build. You can even pass them along to other professors.
So, if molecular cloning is in your future, let us help you get organized. Try the CloneWeaver® Workflow Builder.
Like this:
Like Loading...
 Let’s face it, most lab techs and purchasing agents aren’t all that happy when you send them an Instagram picture of your latest lunchroom-napkin cloning strategy as your order form for your next big cloning experiment. So we have created the
Let’s face it, most lab techs and purchasing agents aren’t all that happy when you send them an Instagram picture of your latest lunchroom-napkin cloning strategy as your order form for your next big cloning experiment. So we have created the  Already have a favorite vector and a freezer full of restriction enzymes? No problem, skip those steps and move on to getting the perfectly sized nucleic acid markers or the particular polymerase your experiment requires.
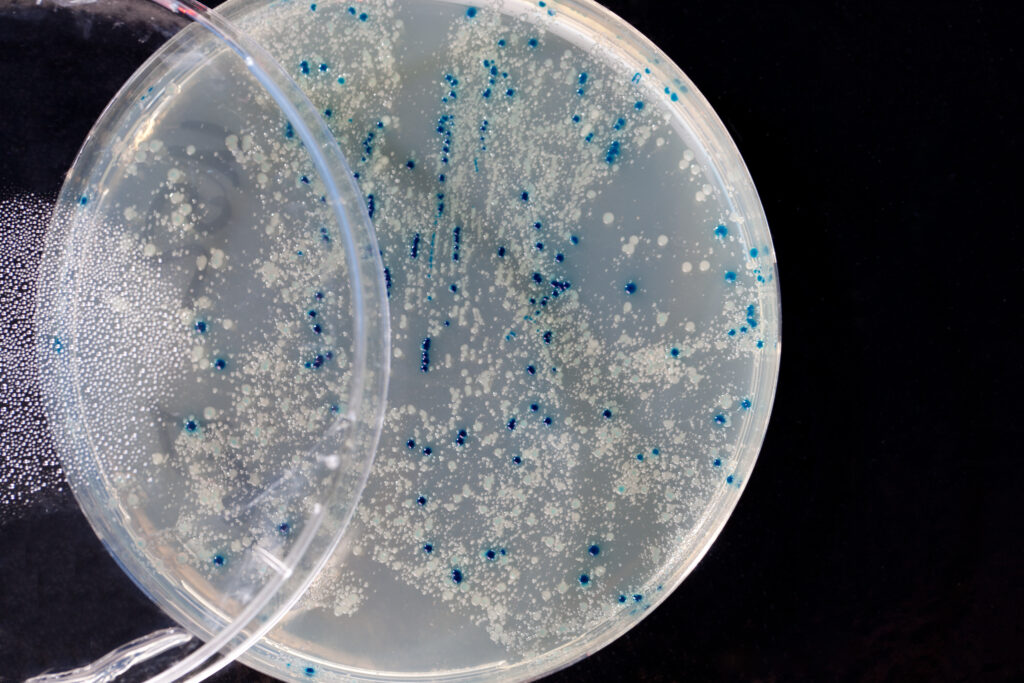
Already have a favorite vector and a freezer full of restriction enzymes? No problem, skip those steps and move on to getting the perfectly sized nucleic acid markers or the particular polymerase your experiment requires.![4498MA-[Converted]](https://www.promegaconnections.com/wp-content/uploads/2014/10/4498MA-Converted-e1413386878386.jpg)