This is the second in a series of four blogs about Quantitation for NGS is written by guest blogger Adam Blatter, Product Specialist in Integrated Solutions at Promega.
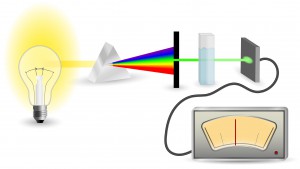
Perhaps the most ubiquitous quantitation method is UV-spectrophotometry (also called absorbance spectroscopy). This technique takes advantage of the Beer-Lambert Law: an observation that many compounds absorb UV-Visible light at unique wavelengths, and that for a fixed path length the absorbance of a solution is directly proportional to the concentration of the absorbing species. DNA, for example has a peak absorbance at 260nm (A260nm).
This method is user friendly, quick and easy. But, it has significant limitations, especially when quantitating samples for NGS applications.
- UV-absorbance lacks sensitivity. Even with a perfectly pure sample, the lowest detectable concentration of DNA is 2ng/µl. When you are performing NGS applications, often you are working with even lower concentrations of nucleic acid.
- Unfortunately, DNA is not the only molecule that absorbs light at 260nm. Many other organic compounds, including proteins, common contaminants left over from purification methods like phenols, and other nucleic acids (RNA, ssDNA, primers), also absorb at 260nm. Because these non-template compounds will contribute to the absorbance reading, they can cause gross overestimation of the the sample concentration.
These limitations and complicating factors can lead to insufficient input for library preparation or uneven coverage in multiplex pools as a result of poor normalization. In some cases, you would not be able to discern results from background; in others the run might fail completely. For these reasons, many NGS assay instruction manuals recommend recommend avoiding UV-absorbance-based quantitation.
Read Part 1: When Every Step Counts: Quantitation for NGS
Read Part 3: Fluorescence Dye-Based Quantitation: Sensitive and Specific for NGS Applications
Read Part 4: Real-Time (Quantitative) qPCR for Quantitating Library Prep before NGS
Latest posts by Promega (see all)
- Accurate and On-Time: A Look Inside Promega Logistics - June 24, 2025
- Augmenting Human Capabilities with AI Tools - June 17, 2025
- Lead with Empathy: Supporting Caregivers in the Workplace - June 3, 2025
