Most of us have experienced an inflammatory response at some point in our lives. Fever, achy joints, swelling around a scrape or cut, all of these are forms of inflammatory response. Inflammation is the body’s response to infection or tissue damage and acts to limit harm to the rest of the body. A key player in the inflammation process is a group of protein complexes call inflammasomes. The term “inflammasome” was first used in 2002 by researchers in Switzerland (1) to refer to a caspase-activating protein complex. We now know that inflammasomes are cytosolic multiprotein platforms that assemble in response to pathogens and other signals. Inflammasome assembly results in the processing of the inactive procaspase-1 into the active cysteine-protease enzyme, caspase-1. Caspase-1, in turn, activates the proinflammatory cytokines Interleukins IL-1β and IL-18. In addition, caspase-1 is also required for pyroptosis, which is an inflammatory form of cell death that combines the characteristics of apoptosis (fragmented DNA) and necrosis (inflammation and cytokine release) and is frequently associated with microbial infections.
Inflammasome complexes are made up of scaffolding sensor proteins (NLR, AIM2, ALR), and an adaptor protein containing a caspase activation and retention domain (CARD) and inactive procaspase-1. Most inflammasomes are formed with one or two NLRs (NOD-like receptors). However, non-NLR proteins such as AIM2 (absent in melanoma 2) and pyrin can also form inflammasomes. The different sensor proteins are activated by different types of outside stimuli, and inflammasomes are loosely sorted into families based on the protein forming these sensors.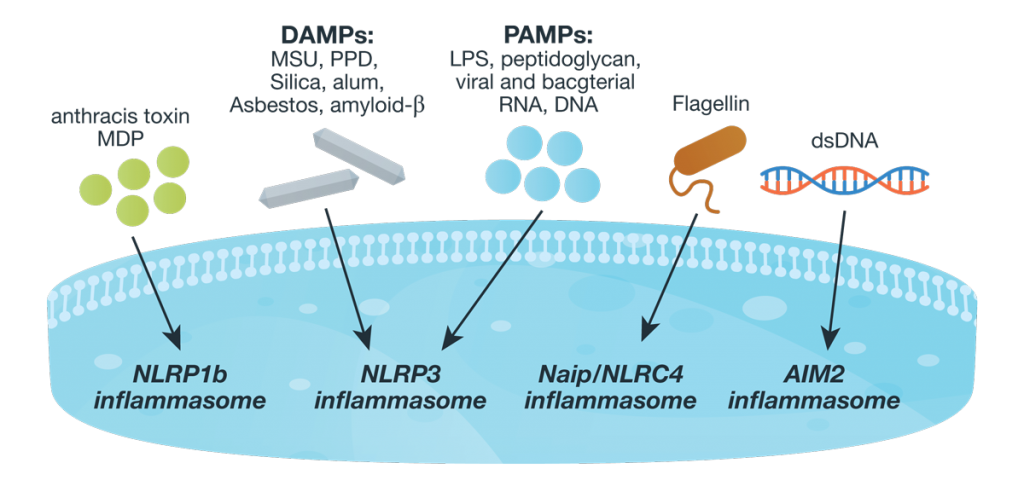
The best characterized family of inflammasomes are those containing NLR sensors. These NLR inflammasomes are activated when the scaffold protein senses or binds its activating stimuli. The NLRP3 inflammasome is the most widely studied inflammasome because it is activated by many different bacterial, viral and fungal pathogens as well as many host-derived factors and crystalline substances (e.g., silica and alum). It is also suspected to play a role in metabolic diseases and sterile inflammation (a systemic inflammation in the absence of known infectious agents that often presents in chronic form and does not respond fully to treatment; 2).
In contrast to the broad range of NLRP3 activators, the NLRP1b inflammasome activation is highly specific. This inflammasome was the first to be described, and is only activated by anthrax lethal toxin (LeTx) and muramyl dipeptide (MDP; 3).
The NLRC4 inflammasome (also referred to as IPAF) is activated by bacterial type III secretion system (T3SS) components and flagellin (in mice), but unlike other inflammasomes, NLRC4 activation requires a second NLR protein, NAIP, which functions as a receptor for the T3SS triggers. The NLRC4 inflammasome also has a differential requirement for the adaptor protein. The pyrin domain (PYD) and CARD-containing adaptor, ACS is required necessary for caspase-1 activation but not needed for NLRCP-mediated pyroptosis (2).
NLRP6 is a functionally unique inflammasome. Activation of NLRP6 results in downstream effects that are not expected in traditional inflammasome activation. Specifically, NRLP6 has been shown to play a crucial role in maintaining intestinal health by regulating the composition of the microbe population in the colon through IL-18 activation. Studies have also shown that NLRP6 deficiency leads to decreased autophagosome formation, resulting in impaired exocytosis of mucin granule from goblet cells. This defective mucus secretion increases the susceptibility to persistent infection. However, the exact mechanism through which this occurs is still unresolved (2).
Studies have shown that NLRP12 inflammasome activation is associated with Yersinia pestis infections, and NLRP12 complexes are present in monocytes of malaria patients. Despite these clear indicators that NLRC12 plays a relevant role in innate immune responses against pathogens, the specific triggers that induce activity have not been determined (2).
Unlike the NLR family, ALR inflammasomes, AIM2 and IFL16, interact directly with their ligands (dsDNA). These inflammasomes do not contain CARD domains and so must recruit ASC using their PYD for activation. The AIM2 inflammasome is activated by the presence of bacterial or viral dsDNA in the cytosol. Although AIM2 does not seem to differentiate the dsDNA source, a minimum length of 80 base pairs is needed for effective activation. In contrast, IFL16, which is located in the nucleus, sense DNA from Kaposis-associated herpes virus (2).
Pyrin is a non-NLR inflammasome that is expressed primarily in immune cells such as monocytes and dendritic cells. Typically, Pyrin is associated with autoinflammatory diseases because mutations in the pyrin-encoding gene, MEFV, causes the hereditary inflammatory disorder, familial Mediterranean fever. In addition, unlike other pattern recognition receptors that recognize pathogen-associated molecular patterns (PAMPs), pryin is activated by pathogen-induced alterations in cellular machinery. Specifically, modifications in Rho GTPases. By sensing these pathogen-induced changes, pyrin acts to induce the appropriate immune response (2).
Inflammasomes are implicated in a number of inflammatory disorders and in the initiation or progression of diseases such as Alzheimer disease, Parkinson disease, multiple sclerosis, metabolic syndrome, atherosclerosis and inflammatory bowel disease (4, 5). Understanding the mechanisms of inflammasome activation will likely prove critical to understanding and treating many diseases where inflammasome activation causes, instead of protects from, harm
References
- Martinon, F. et al. (2002) The inflammasome: A molecular platform triggering activation of inflammatory casases and processing of proIL-beta. Mol. Cell 10, 417–26.
- Vanaja, S.K., et al. (2014) Mechanisms of inflammasome activation: recent advances and novel insights. Trends in Cell Biol. 25, 308–15.
- Schroder, K and Tschopp, J. (2010) The inflammasome. Cell 140, 821–32.
- Strowig, T. et al. (2012) Infalmmasomes in health and disease. Nature 481, 278–86.
- Gio, H. (2015) Inflammasomes: mechanism of action, role in disease and therapeutics. Nat. Med. 21, 677–87.
Kelly Grooms
Latest posts by Kelly Grooms (see all)
- The Battle of Shiloh’s Angel’s Glow: Fact, Civil War Legend or Modern Myth? - July 11, 2024
- Mind Control, Mutilation and Death. The Fungal Fate That Lurks in Waiting for Emerging Periodical Cicadas - June 13, 2024
- Measles and Immunosuppression—When Getting Well Means You Can Still Get Sick - May 13, 2024
