Targeted protein degradation (TPD) is an emerging drug discovery strategy that offers an entirely different approach to tackle disease-relevant proteins, including classic “undruggable” targets. Instead of inhibiting protein function, small molecules like PROTACs and molecular glues co-opt the cell’s own ubiquitin-proteasome system to eliminate specific proteins altogether. But as this targeted approach gains traction, it also challenges existing methods for validating compound activity.
How do you confirm that degradation is happening in a biologically-relevant system? Can you validate protein degradation in real-time?
Bioluminescence offers a powerful solution to both track and quantify protein degradation in real time. In this blog, we explore how bioluminescent assays can be combined with live-cell imaging on the GloMax® Galaxy Bioluminescence Imager to provide a more complete view of protein degradation in real time. Using GSPT1 as a model target, we show how this workflow helps confirm degradation activity and visualize degradation over time, directly in live cells.
Tracking Targeted Protein Degradation with HiBiT Reporters
Before studying how a degrader compound affects protein abundance, researchers need to ensure they’re working with an appropriate cellular model. Some groups have adopted cell-free systems or engineered reporter lines that provide a convenient and chemically tractable way to screen degrader activity. While these cell-free systems may reflect the intended chemistry, they don’t always reflect the native cellular environment1. Other “model” systems attempt to bridge this gap by inserting plasmids into easily manipulated cell lines (like E. coli or Pseudomonus), but come with their own limitations like overexpression artifacts, lack of cell-type specificity, or restricted compatibility with primary or disease-relevant cells2.
To address these limitations and bring degradation studies into more physiologically relevant context, researchers are turning to tools that more accurately match endogenous protein activity. The HiBiT system offers a powerful way to monitor protein levels in live cells, with high sensitivity and minimal disruption. HiBiT is an 11–amino acid peptide tag that can be inserted into endogenous proteins using CRISPR/Cas9 gene editing, eliminating the problems surrounding plasmid overexpression or artificial reporter constructs.
Once expressed in cells, the HiBiT-tagged protein can be detected by adding LgBiT, a larger protein fragment that complements HiBiT with high-affinity to form a fully functional luciferase enzyme (Figure 1). In the presence of a substrate, a bright luminescent signal is generated proportional to the amount of target protein present.
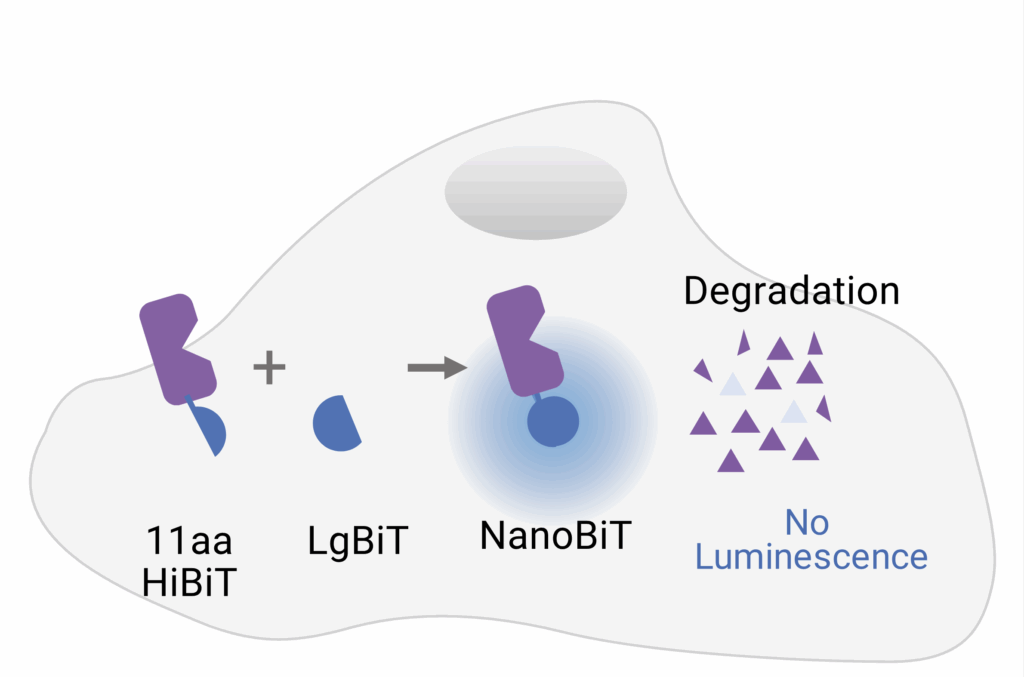
This luminescent signal has traditionally been measured using plate-based luminometers, such as the GloMax® Discover. While these systems are robust and quantitative, they provide only population-level data and can fail to discern cell-to-cell differences. Imaging the bioluminescence generated by HiBiT adds a new layer of insight. The GloMax® Galaxy Bioluminescence Imager was designed for imaging NanoLuc® Luciferase based technologies, such as HiBiT, and offers validation by capturing images of what’s actually occurring at the cellular level. With the ability to image HiBiT signal in live cells using the same detection chemistry as plate-based readers, it bridges the gap between quantification and confirmation. For researchers developing degrader compounds or screening CRISPR knock-ins, this live-cell imaging step helps confirm that the biology they’re studying is the biology they intended to model.
Case Study: Visualizing GSPT1 Degradation in Live Cells
To illustrate how luminescent imaging strengthens protein degradation workflows, researchers at Promega used HiBiT-based assays to monitor the degradation of GSPT1, a translation termination factor essential for cell proliferation and survival. GSPT1 has been implicated in leukemia and other cancers, making it a valuable target for therapeutic intervention. Instead of blocking GSPT1 activity directly, the researchers used a molecular glue (CC-885) to signal degradation by controlling the native cellular degradation machinery. Specifically, CC-885 binds to cereblon (CRBN), a substrate receptor within the native ubiquitin-ligase complex. This binding event reshapes the surface or CRBN, enabling it to recruit new protein targets that would otherwise be ignored. Once recruited, these targets (like GSPT1) are tagged with ubiquitin molecules and directed to the proteasome, a large protein complex that acts as the garbage disposal of the cell.
In this study, researchers used plate-based luminometer assays to demonstrate that increasing doses of CC-885 led to a clear, dose-dependent decrease in luminescent signal from HiBiT-GSPT1 cells, indicating robust degradation activity. But since this signal represents an average across thousands of cells, the team supplemented this work with bioluminescent imaging. Live-cell imaging revealed a consistent reduction in luminescence after treatment with 100nM CC-885, confirming that GSPT1 degradation was widespread across the population and sustained over time. In contrast, control groups showed only a gradual decline in signal, consistent with normal HiBiT turnover. Brightfield imaging confirmed that cellular structure remained largely intact, although a slight curling of the cell edges in treated wells suggested early signs of cytotoxicity. Together, these findings show how bioluminescent imaging compliments targeted protein degradation studies, enabling researchers to visualize degradation in real-time.
To learn more about the experiment, see the full application note here.
Interested in learning more about bioluminescence imaging with the GloMax® Galaxy System? Explore our Applications Hub, where we dive into the value added through imaging your NanoLuc® Luciferase assays. The GloMax® Galaxy Bioluminescence Imager is for research use only.
References
- Brio, L., et al. “Affinity Microfluidics Enables High-Throughput Protein Degradation Analysis in Cell-Free Extracts.” Communications Biology, vol. 5, 2022, article 1147. Nature, doi:10.1038/s42003-022-04103-3. ↩︎
- James, J., et al. “Protein Over-Expression in Escherichia coli Triggers Adaptation Analogous to Antimicrobial Resistance.” Microbial Cell Factories, vol. 20, 2021, article 13. BioMed Central, doi:10.1186/s12934-020-01462-6. ↩︎
Anna Bennett
Latest posts by Anna Bennett (see all)
- A New View of Protein Degradation with HiBiT and Live Cell Imaging - September 23, 2025
- HiBiT-Based NanoBRET® Assay Sheds Light on GPCR–Ligand Binding in Live Cells - July 31, 2025
- Understanding Wnt Signaling Through β-Catenin Localization in Live Cells - June 19, 2025
