As the SARS‐CoV‐2 pandemic continues to rage across the United States and around the globe, the demand for COVID‐19 testing is increasing. The vast majority of the COVID-19 assays use RT‐qPCR to detect the viral RNA in patient samples such as nasopharyngeal swabs, which are collected and stored in viral or universal transport media (VTM/UTM). The general workflow for these COVID‐19 assays can be broken down as follows:
- Collect and store patient samples
- Ship samples to testing laboratory
- Extract RNA from samples
- Amplify and analyze samples
While many companies who manufacture the products that are used in these steps have been able to adapt and significantly increase their production capacities, there are still gaps between the supply of these products and the global test demand. Both the sample collection and storage step and the RNA extraction/purification step have a tendency to bottleneck and experience supply constraints. One way to address these bottlenecks and expand production capacity for these in‐demand products is to evaluate the viability of skipping a step in the workflow, without hindering the ability to detect viral RNA from samples.
Though we manufacture and supply RNA extraction products here at Promega, we recognized RNA extraction as a particularly rate‐limiting step in the current COVID‐19 testing workflow. Leveraging our 40+ years of experience in both sample preparation and sample amplification, especially our work with challenging samples such as forensic crime scene evidence, we sought a solution that would allow the analysis of nasopharyngeal swabs stored in VTM/UTM without first performing RNA extraction. The result is a direct PCR amplification method that simplifies and accelerates time to qPCR results.
As the name implies, direct PCR is a method that enables direct PCR amplification of a sample without prior RNA or DNA extraction and purification steps. By removing those more time‐consuming steps, you can complete the upstream process more quickly, enabling you to move on to the downstream processing faster.
Challenges in Direct Amplification
Direct PCR amplification comes with its own set of challenges. As with any PCR assay, PCR inhibition is always a factor for consideration. PCR inhibitors—from both the sample and the lysis buffer utilized to lyse and release the RNA—have the potential to skew quantification in qPCR. In a typical PCR workflow, the solution to PCR inhibition is to purify or dilute the RNA extracts, which leads to a loss of RNA. Since direct amplification skips the purification step, it is helpful to have an understanding of underlying molecular inhibition mechanisms to help overcome any analytical limitations that you may experience in the presence of PCR inhibitors.
The second main challenge to overcome in direct PCR amplification is assay sensitivity. Prior to analysis, RNA extraction can serve as a concentrating step, usually beginning with Xµl of VTM and eluting in Yµl—resulting in a Z‐fold increase in the sample concentration going into the RT‐qPCR assay. In direct amplification, there is no sample concentration. In fact, you are usually going to dilute the sample during the lysis step to release the RNA. To overcome this, you many need to re‐optimize your PCR assay, given that a PCR assay that is optimized to amplify extracted RNA may not be optimized for a direct‐from‐sample amplification.
Benefits of Direct Amplification
Despite its challenges, there are many benefits to direct PCR amplification. In a typical PCR workflow, the extraction and purification wash steps utilize many buffers and optimization reagents. In a direct amplification workflow, you are able to circumvent these costly and rate‐limiting steps, allowing you to avoid potential throughput limitations from reagent supply constraint and enabling you to cut costs and conserve precious sample material.
Utilizing a direct PCR workflow additionally helps save you time in the lab, though the amount of time savings will vary depending on if your previous RNA extraction method was automated or manual. Direct amplification workflows are also automation‐friendly, requiring no dependency on specially‐ designed plastic consumables.
Our Solution: XpressAmp™ Direct Amplification Reagents
The XpressAmp™ Direct Amplification Reagents provide a fast, RNA extraction‐free method to prepare viral samples for PCR‐based amplification. Collect the samples by nasopharyngeal swab or in universal or viral transport media, and perform direct amplification analysis using RT‐qPCR. The simple sample preparation method requires only a 10‐minute, room temperature incubation that is easily automated for high‐throughput needs.
💥New! Protocol for Direct Amplification of Viral RNA from Saliva. Download the PDF protocol.
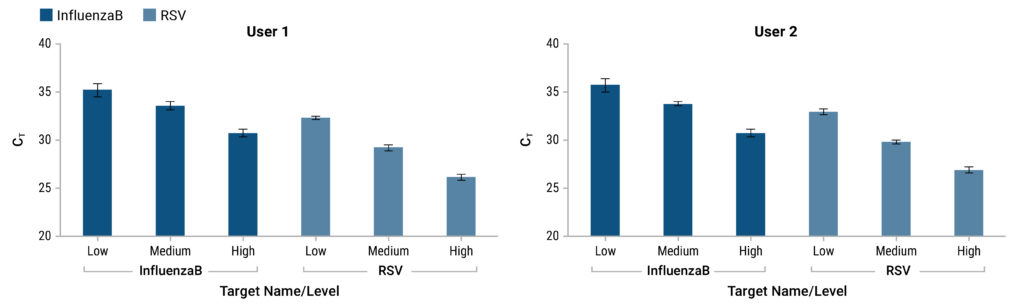
Detection of Viral RNA Without the Need for RNA Extraction
Simple, Automation-Friendly Protocol

Customizable
The XpressAmp™ Direct Amplification Reagents can be adjusted in numerous ways to suit your specific needs. Many customization options are available to you, from purchasing the reagents in bulk, to changing the dispense size, or reformatting and relabeling.
Learn more about the XpressAmp™ Direct Amplification Reagents and the ability to customize to your needs.
Want to know more about how the Maxwell® RSC can give you the freedom to focus on the work that interests you the most? To learn more, click here.
