A total of five human remains were used for this study. Four individuals were found in a mass grave under the sacristy of a Catholic church in southern Germany. These remains were radiocarbon dated to the 14th century. The fifth individual was a soldier unearthed with from a grave with two other soldiers in Brandenburg located in northeastern Germany. The soldiers’ remains were dated to the Thirty Years’ War, which occurred 1618–1648. Seifert et al. genotyped the ancient Y. pestis DNA using 16 short nucleotide polymorphisms (SNPs) based on the likely phylogenetic location of the recovered strains. One SNP confirmed whether the tested strain differed from the ancestral branch. A second SNP determined that the strains did not differ into another branch separate from currently typed Black Death strains. The remaining 12 SNPs assessed how closely or distantly related German Y. pestis strains were to the currently known Black Death variants.
Interestingly, the Y. pestis samples typed from five German remains were identical for all 16 SNPs. Seifert et al. suggest this may mean that there could be a local reservoir of Y. pestis that may be reinfecting the population rather than being reintroduced from Asia, a prevailing hypothesis. However, this observation raises more questions. What was the geographic origin of the soldier in Brandenburg? That is, did he come from southern Germany to fight in the Thirty Years’ War or was he local to northeastern Germany? Could there be a nearby Asian source of this SNP genotype that continued to reinfect the German population over 300 years?
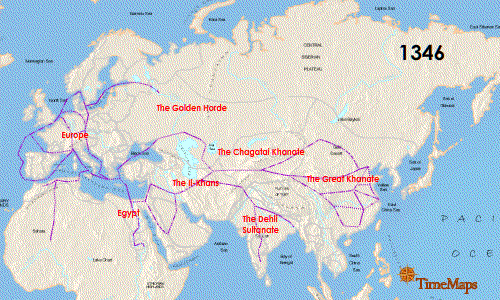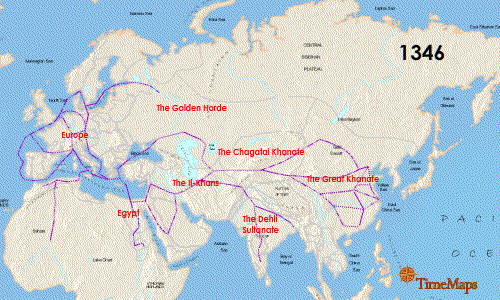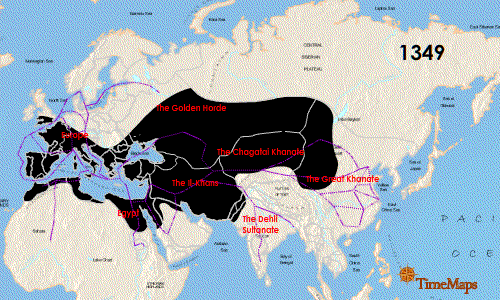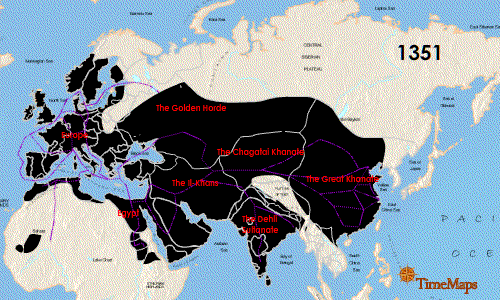
Researchers also noted that the German genotype was similar to that found in East Smithfield cemetery in London, England with an overlap of 13 identical SNPs as well as a Hereford, England isolate (4 matching SNPs) and a French isolate (3 matching SNPs). While there were similarities especially with the London sample, the use of 16 SNPs means there are missing matches that could be made to strengthen the association between the English and German genotypes because the East Smithfield sample was typed with 22 SNPs and lacked 2 SNPs present in the German analysis. If additional genotyping more closely matches the East Smithfield Y. pestis SNP genotype and those of other Black Death remains, what does this mean for the possible local German source of the genotype? Could this just be a successful Y. pestis variant that persisted for the entire 300 years? Finding more infected remains over this same time frame in Europe and Asia could help determine if there was a potential European source of the genotype or if it persisted in Asia and was just reintroduced repeatedly.
Seifert et al. were able to genotype German remains separated by time and location and discovered an identical 16 SNP genotype. Could this mean there was a localized source of Y. pestis strain that continued to reinfect that part of Europe? That is a possibility, but the researchers acknowledged more SNP genotyping and whole genome sequencing of other plague victims could increase understanding of the strains associated with the Black Death and the full 300-year span of the second pandemic. I would like to see data from Asian remains as well to help fill in the picture of the breadth of Y. pestis strains infecting humans during this period.
Reference
Seifert, L. et al. (2016) Genotyping Yersinia pestis in historical plague: Evidence for long-term persistence of Y. pestis in Europe from the 14th to the 17th century. PLOS ONE 11, e0145194. doi: 10.1371/journal.pone.0145194
Sara Klink
Latest posts by Sara Klink (see all)
- A One-Two Punch to Knock Out HIV - September 28, 2021
- Toxicity Studies in Organoid Models: Developing an Alternative to Animal Testing - June 10, 2021
- Herd Immunity: What the Flock Are You Talking About? - May 10, 2021

One thoughtful comment