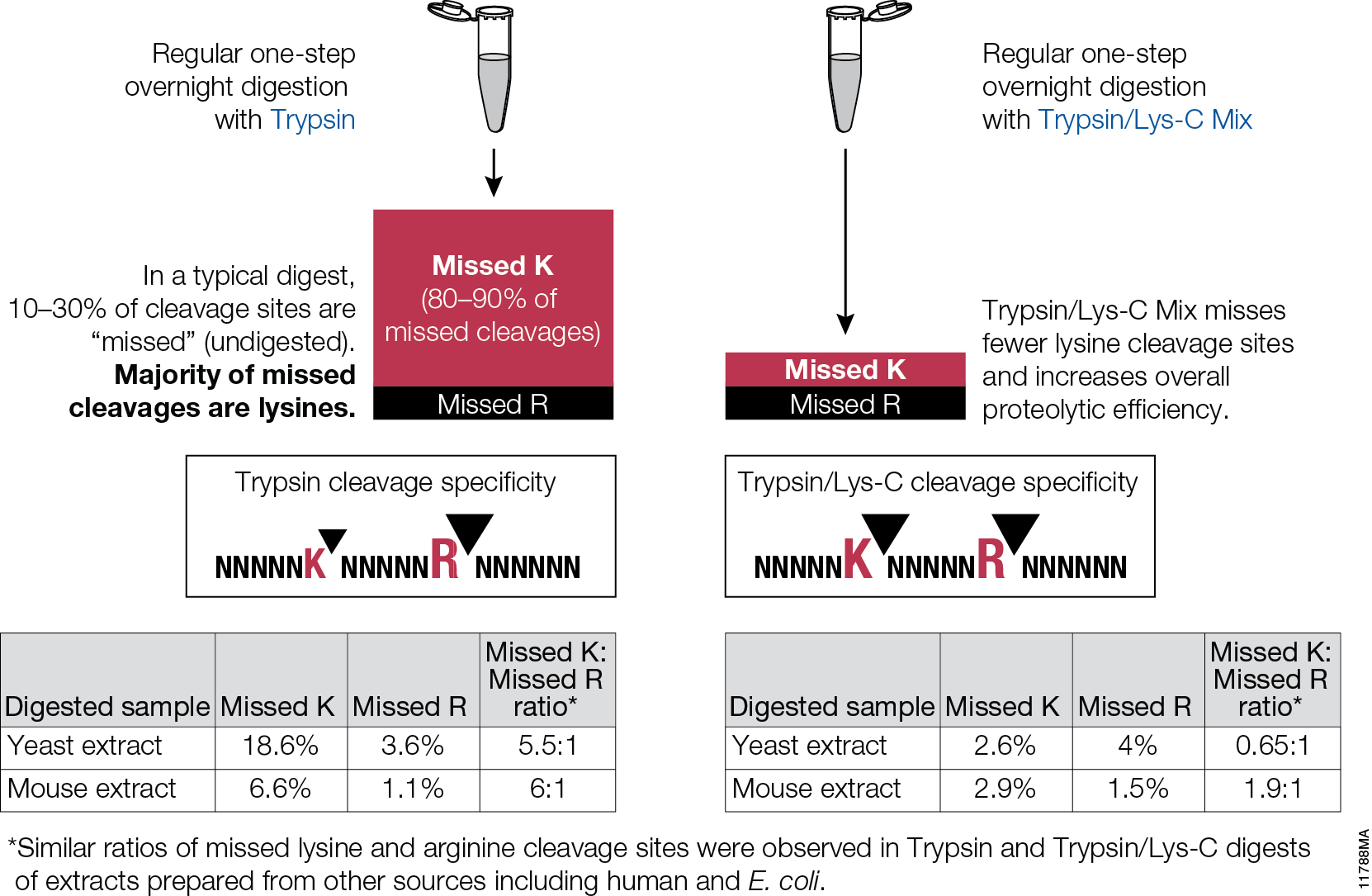
It’s time to analyze your protein and you are trying to decide where to begin. You are asking questions like: Which protease do I choose? How much enzyme should I use in my digest? How long should I perform my digest?
Unfortunately, there is no one-size fits all answer to this type of question other than… “well it depends.” All protease digests will be a balance between denaturing the protein sample to allow access to cleavage sites, optimizing conditions for the protease to function, and compatibility with your workflow and downstream applications. We provide general guidelines that work for most samples, but frequently you will need to optimize the conditions need for your specific sample and application.
Here, I use the example of a trypsin digest for downstream mass spectrometry to highlight key questions to ask and factors that can be optimized for any digest.
Continue reading “What’s In YOUR Protein? Optimizing Protease Digestions to Get the Inside Scoop”