The SARS-CoV-2 nucleocapsid protein accounts for the largest proportion of viral structural proteins and is the most abundant protein in infected cells. Nucleocapsid proteins have the job of “packaging” the viral nucleic acid (in this case, RNA). Viral nucleocapsid proteins can also enter the host nucleus and interact with a variety of host proteins to interfere with critical processes of the host cell, including protein degradation. Here we review a study that used an in vitro protein degradation assay to investigate the interaction of the SARS-CoV-2 nucleocapsid protein and the proteasome activator PA28γ.
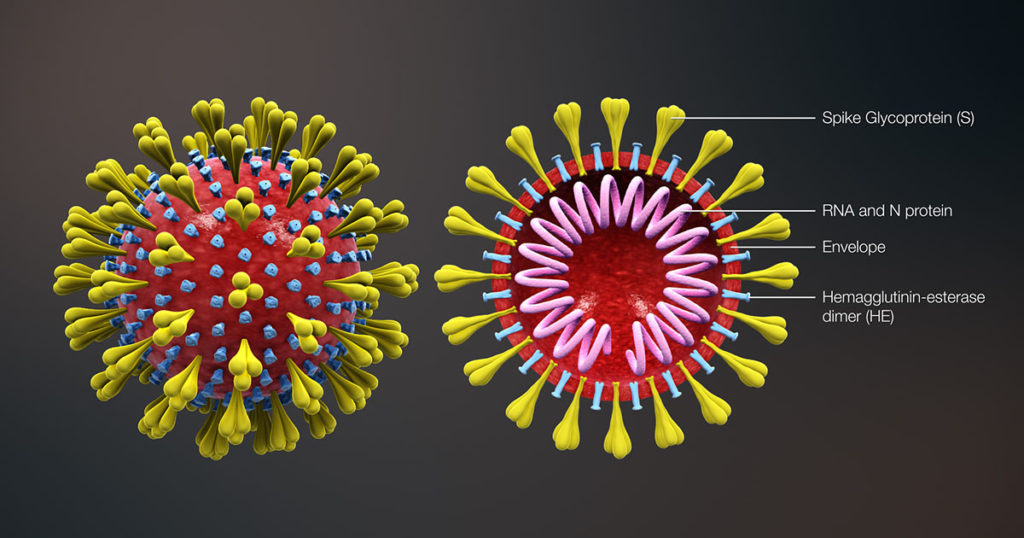
In SARS-CoV-2 infections, the nucleocapsid protein is critical for infection, replication and packaging. The SARS-CoV-2 nucleocapsid protein is not only localized in the cytosol of the host cell but also is translocated into the nucleus. There, it interacts with various cellular proteins that modulate cellular functions, such as the degradation of unneeded or damaged proteins by proteolysis. Researchers have proposed that the protein degradation system plays an important part in coronavirus infection (1).
Continue reading “SARS-CoV-2 Nucleocapsid Protein and PA28γ: A Role in Pathogenesis?”