In a study published in Proceedings of the National Academy of Sciences USA article, Wang et al. used the principle of the Promega NanoBRET™ assay to understand how ERK1/2 phosphorylation of Rabin8, a guanine nucleotide exchange factor, influenced its configuration and subsequent activation of Rab8, a protein that regulates exocytosis.
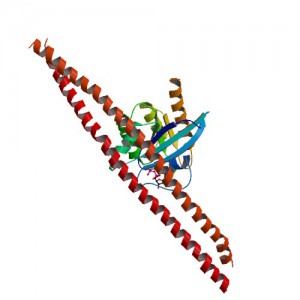
Rab8 is a member of the Rab family of small GTPases and an important regulator of membrane trafficking from the trans Golgi network and recycling endosomes to the plasma membrane. Wang et al. were interested in learning how the guanine nucleotide exchange factor (GEF) Rabin8, a known activator of Rab8, was itself activated to better understand how Rab8 and exocytosis were regulated in the cell. First, they confirmed if the consensus extracellular-signal-regulated kinases ERK1/2 phosphorylation motif uncovered in Rabin8 resulted in phosphorylation of Rabin8. Both in vitro analysis and cell-based assays confirmed that ERK1/2 phosphorylated Rabin8. Next, the GEF activity of Rabin8 was assessed to determine if ERK1/2 phosphorylation activated the GEF. Researchers confirmed activation of Rabin8 GEF in vitro.
Continue reading “Uncovering Protein Autoinhibition Using NanoBRET™ Technology”