On July 3rd and 4th, 2021, we celebrate World Firefly Day, and 2021, marks 30 years of luciferase products firefly luciferase vectors and Luciferase Assay System. These tools are key in advancing bioluminescent technology. To celebrate this day, we want to highlight some innovations that have been made possible with these tools.
Continue reading “World Firefly Day: Shining More Light on Glo-ing Innovations”bioluminescent assay
Drug Repurposing Screens: Redeploying Old Dogs for New Tricks
This blog was written by guest author, Amy Landreman, PhD.
Drug repurposing, identifying new uses for approved or investigational drugs, is an attractive strategy when looking for new disease treatments. Because the compounds have already gone through some level of pre-clinical optimization and safety testing, this approach can reduce risk, reduce cost, and speed up the timeline for further drug development. An additional benefit of this approach is that it can result in new biological insights or a better understanding of disease mechanisms since these compounds usually already have some level of mechanistic characterization. Indeed, there are now a number of compound collections openly available specifically for the purpose of facilitating drug repurposing efforts. For example, the ReFRAME (Repurposing, Focused Rescue, and Accelerated Medchem) library is a collection of 12,000 compounds developed by Scripps Research Center and has been screened to identify novel candidate therapeutics for Cryptosporidium infection (1). The Broad Institute also offers a drug repurposing hub that contains an annotated collection of over 7,000 compounds.

Drug repurposing libraries, although often smaller than novel compound small molecule libraries, are designed for implementation into high-throughput screening workflows in order to efficiently triage compounds for the desired result. Effective compound screens require assays that can be scaled to 384 or 1536-well microplate formats and implemented in batch or continuous processing workflows. The firefly luciferase reaction has been leveraged to create many assays that are well-suited to these types of high-throughput screening approaches. In particular, the generation of “Glow” assays that have stable luminescent signals and homogenous assay design is a good fit. The signal stability allows for multi-plate processing and because the reagent is added directly to cells in culture, pre-processing steps are eliminated allowing for automated workflows. Assay reagents such as the CellTiter-Glo® Cell Viability Assay and the ADP-Glo™ Kinase Assay are commonly used in screening efforts including those done with repurposing libraries. In addition, there are several firefly luciferase reporter assay reagents such as Steady-Glo® and Bright-Glo™ Luciferase Assays that have been optimized for high-throughput detection of firefly luciferase activity making them well-suited to repurposing screens.
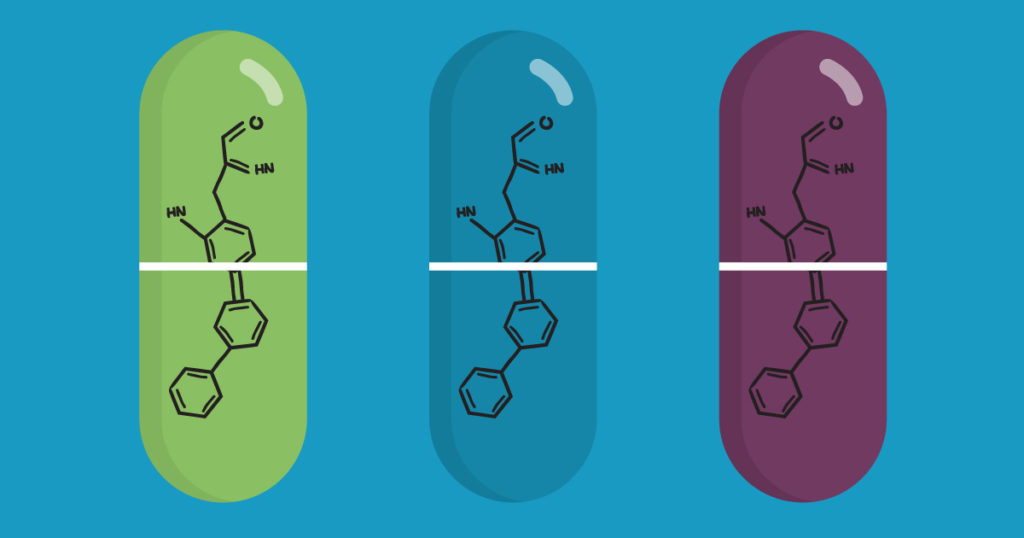
Firefly Luciferase Sheds Light on Development of New Malaria Treatments

Despite significant advancements in antimalarial drugs and widespread efforts to prevent transmission over the past decade, deaths from malaria remain high, particularly in younger children. New drugs with novel modes of action are urgently needed to continue reducing mortality and address drug resistance in the malaria parasite, Plasmodium falciparum. While tens of thousands of compounds have been identified as potential candidates through massive screening efforts, scalable methods for identifying the most effective compounds are needed.
The goal is to find a drug that is potent during all stages in the life cycle of P. falciparum and kills the parasite quickly. Focusing on assessing whether a compound can rapidly eliminate initial parasite burden, Paul Horrocks, PhD, and his colleagues developed a validated bioluminescence-based assay that rapidly determines the initial rate of kill for discovery antimalarials. One key to developing their assay was figuring out how to monitor when the parasite dies after introducing the drug. While measuring DNA content can be used to monitor parasite burden, it is too stable to use for a relevant time course assay.
Enter firefly luciferase, a dynamic reporter tool to investigate drug action. By creating transgenic P. falciparum that express the luc reporter gene, the researchers could monitor drug action over time. When the parasite is killed, it stops making the luciferase reporter. Since there is no new production of luciferase, levels fall quickly after the parasite dies, and a luciferase assay can determine how fast each drug killed the parasite.
Continue reading “Firefly Luciferase Sheds Light on Development of New Malaria Treatments”Celebrating 30 Years of “Glo-ing” Research
This post is written by guest blogger, Amy Landreman, PhD, Sr. Product Manger at Promega Corporation.
In December of 1990, Promega first discussed the use of firefly luciferase (luc) as an emerging reporter technology in the article, Firefly Luciferase: A New Tool for Molecular Biologists. At the time, the gene coding chloramphenicol acetyltransferase (cat) was most commonly used by researchers, but it was thought that the bioluminescent properties of firefly luciferase, extreme sensitivity and rapid simple detection, could make a significant difference in how molecular biologists tackled their research. Several months later, the first firefly luciferase reporter vectors and detection reagents became available as products, making this new technology more broadly accessible to the research community. Today firefly luciferase is no longer a “new tool”, with it and many other bioluminescent reporter technologies being standard elements of the modern research toolbox.

A Cell Viability Assay for Today

Cell viability assays are a bread-and-butter method for many researchers using cultured cells —everyday lab tools that are a part of many newsworthy papers, but rarely make news themselves.
Over time, cell viability assays have become easier to use and more “plug ‘n play”. Among modern assays, luminescent plate-reader based systems have been a favorite for several years because of their superior sensitivity, robustness, simple protocols and uncomplicated equipment requirements (all you need is a plate-reading luminometer). These qualities combine to allow easy scalability and adaptability from bench research to high throughput applications.
CellTiter-Glo® Luminescent Cell Viability Assay is an accepted go-to viability assay for many researchers. The assay measures ATP as an indicator of metabolically active cells. A quick search on Google Scholar returns 3,990 CellTiter-Glo results for 2017 and over 500 so far in January and February of 2018. A sampling of these recent publications gives a snapshot of some of the ways the CellTiter-Glo assay is used to support key areas of research today.
Does a treatment kill cells?
The obvious application of a cell viability assay is to understand whether cells are alive. In cancer research, the CellTiter-Glo assay is often used to confirm killing of tumor cells and to verify that normal cells survive. Therefore, these assays are a key part of the evaluation and screening of drug candidates and other therapies for cancer. Many papers reporting use of CellTiter-Glo are developing and evaluating the effectiveness of novel anti-cancer treatments. Continue reading “A Cell Viability Assay for Today”
Screening for Antiviral Compounds under Level 4 Containment Conditions
Working with bacteria and viruses that cause life-threatening diseases with no currently available treatment options takes guts. Most scientists are familiar with the routine requirements of good aseptic technique, are highly aware of laboratory safety requirements, and are more than familiar with autoclaves and sterilization issues, but if we make a mistake the consequences are usually only lost time or a spoiled experiment—not a lost life.
Scientists working with highly virulent organisms deal with a whole other level of risk that requires adherence to the strictest of safety regulations, and these containment regulations can sometimes place constraints on the type of experiment that can be performed with dangerous pathogens. A paper published in the April 2014 issue of Assay and Drug Development Technologies brought this to my attention and reminded me of the serious issues some scientists face on a daily basis as they research ways to combat infectious diseases.
Continue reading “Screening for Antiviral Compounds under Level 4 Containment Conditions”Top 10 Innovator—Two Years Running
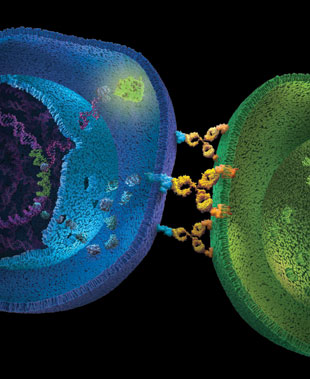
Antibody-dependent cell-mediated cytotoxicity (ADCC) is the main mechanism of action (MOA) of antibodies through which virus-infected or other diseased cells are targeted for destruction by components of the cell-mediated immune system. ADCC assays are often used to assess the effectiveness of monoclonal antibody therapies during the manufacture and development of biologic drugs. The bioluminescent ADCC Reporter Bioassays use an alternative readout at an earlier point in ADCC MOA pathway for the quantification of Fc effector function of antibody-based molecules: the activation of gene transcription through the NFAT (nuclear factor of activated T-cells) pathway in the effector cell.
The bioassay uses engineered Jurkat cells stably expressing the FcγRIIIa receptor, V158 (high affinity) variant, and an NFAT response element driving expression of firefly luciferase. The assay is ADCC MOA-based and features frozen, thaw-and-use effector cells and optimized reagents and protocol to perform a reporter-based ADCC bioassay in a single day. The ADCC Reporter Bioassay correlates with classic cytotoxic ADCC assays and is a suitable replacement for these cumbersome and highly variable assays.
The novel bioassay is linear, accurate, precise and stability indicating. Moreover, the bioassay shows good linear correlation between levels of glycosylation or fucosylation and ADCC activity. All of these features indicate the assay is suitable for use across biologic drug development programs.
Resources for the ADCC Reporter Bioassays:
-
Low Variability ADCC Bioassay
- ADCC Reporter Bioassay, Core Kit Technical Manual
- ADCC Reporter Bioassay, Complete Kit (WIL2-S) Technical Manual
- ADCC Reporter Bioassay Complete Kit (Raji) Technical Manual
- ADCC Reporter Bioassay Effector Cells, Propagation Model Technical Manual
- ADCC Reporter Bioassay: A Novel, Bioluminescent Cell-Based Assay for Quantifying Fc Effector Function of Antibodies (webinar)
