We offer a wide array of GoTaq® DNA Polymerases, Buffers and Master Mixes, so we frequently answer questions about which product would best suit a researcher’s needs. On the Taq Polymerase Page, you can filter the products by clicking the categories on the left hand side of the page to narrow down your search. Here are some guidelines to help you select the match that will best suit your PCR application. Continue reading “How Do I Choose the Right GoTaq® Product to Suit My Needs for EndPoint PCR?”
We offer a wide array of GoTaq® DNA Polymerases, Buffers and Master Mixes, so we frequently answer questions about which product would best suit a researcher’s needs. On the Taq Polymerase Page, you can filter the products by clicking the categories on the left hand side of the page to narrow down your search. Here are some guidelines to help you select the match that will best suit your PCR application. Continue reading “How Do I Choose the Right GoTaq® Product to Suit My Needs for EndPoint PCR?”
PCR, qPCR, and RT-qPCR
Top 10 Tips to Improve Your qPCR or RT-qPCR Assays
Scientists have had a love-hate relationship with PCR amplification for decades. Real-time or quantitative PCR (qPCR) can be an amazingly powerful tool, but just like traditional PCR, it can be quite frustrating. There are several parameters that can influence the success of your PCR assay. We’ve highlighted ten things to consider when trying to improve your qPCR results.
Continue reading “Top 10 Tips to Improve Your qPCR or RT-qPCR Assays”PCR Cloning: Answers to Some Frequently Asked Questions
 Q: What is the easiest way to clone PCR Products?
Q: What is the easiest way to clone PCR Products?
A: The simplest way to clone PCR Products is to amplify the product using thermostable polymerases such as Taq, Tfl or Tth polymerase. These polymerases add a single deoxyadenosine to the 3´-end of the amplified products (3´-end overhang), and can be cloned directly into a linearized T-vector.
Q: What if my DNA polymerase has 3´ to 5´ exonuclease activity (i.e., proofreading activity) that removes the 3´-end overhang?
A: To clone PCR products that have been amplified with a polymerase that have proof reading activity into a T-vector, you will need to perform an A-tailing step using Taq DNA polymerase and dATP. Blunt ended restriction digest fragments can also be A-tailed using this method. The method below uses GoTaq Flexi DNA Polymerase (comes with a Mg-free reaction buffer), but any Taq DNA polymerase can be used.
Set up the following reaction in a thin-walled PCR tube:
1–4µl purified blunt-ended DNA fragment (from PCR or restriction enzyme digestion)
2µl of 5X GoTaq Reaction Buffer (Colorless or Green)
2µl of 1mM dATP (0.2mM final concentration)
1µl GoTaq Flexi DNA Polymerase (5u/µl)
0.6µl of 25mM MgCl2 (1.5mM final concentration)
Nuclease-free water to a final volume of 10µl
Incubate at 70°C for 15–30 minutes in a water bath or thermal cycler.
Q: What is a T-vector, and why are they used for cloning PCR products?
A: T vectors are linearized plasmids that have been treated to add T overhangs to match the A overhangs of the PCR product. PCR fragments that contain an A overhang can be directly ligated to these T-tailed plasmid vectors with no need for further enzymatic treatment other than the action of T4 DNA ligase.
For a complete PCR Cloning protocol, Visit the Cloning Chapter of the Promega Protocols and Applications Guide.
Dealing with PCR Inhibitors
The polymerase chain reaction (PCR) has revolutionized modern biology as a quick and easy way to generate amazing amounts of genomic data. However, when PCR doesn’t work, it can be frustrating. At these times, PCR and reverse transcription PCR (RT-PCR) inhibitors seem to be everywhere: They lie dormant in your starting material and can co-purify with the template of interest, and they can be introduced during sample handling or reaction setup. The effects of these inhibitors can range from partial inhibition and underestimation of the target nucleic acid amount to complete amplification failure. What is a scientist to do?
Continue reading “Dealing with PCR Inhibitors”Methods for Quantitating Your Nucleic Acid Sample
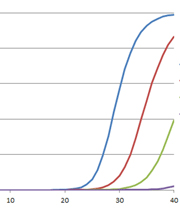
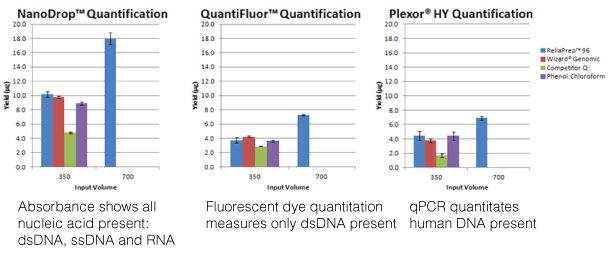
For most molecular biology applications, knowing the amount of nucleic acid present in your purified sample is important. However, one quantitation method might serve better than another, depending on your situation, or you may need to weigh the benefits of a second method to assess the information from the first. Our webinar “To NanoDrop® or Not to NanoDrop®: Choosing the Most Appropriate Method for Nucleic Acid Quantitation” given by Doug Wieczorek, one of our Applications Scientists, discussed three methods for quantitating nucleic acid and outlined their strengths and weaknesses.
Continue reading “Methods for Quantitating Your Nucleic Acid Sample”Top Ten Tips for Successful PCR
We decided to revisit a popular blog from our Promega Connections past for those of you in the amplification world. Enjoy:

-
- Modify reaction buffer composition to adjust pH and salt concentration.
- Titrate the amount of DNA polymerase.
- Add PCR enhancers such as BSA, betaine, DMSO, nonionic detergents, formamide or (NH4)2SO4.
- Switch to hot-start PCR.
- Optimize cycle number and cycling parameters, including denaturation and extension times.
- Choose PCR primer sequences wisely.
- Determine optimal DNA template quantity.
- Clean up your DNA template to remove PCR inhibitors.
- Determine the optimal annealing temperature of your PCR primer pair.
[Drum roll please]…and the most important thing you can do to improve your PCR results is:
- Titrate the magnesium concentration.
Choosing the Right Reverse Transcriptase for Your Project

There are a lot of choices when it comes to reverse transcriptases. Choosing the correct one for your cDNA synthesis and RT-PCR project is important. Here are a few questions that will lead you to right RT for your application: Continue reading “Choosing the Right Reverse Transcriptase for Your Project”
Top Ten Things You Can Do to Improve Your PCR Results

10. Modify reaction buffer composition to adjust pH and salt concentration.
Continue reading “Top Ten Things You Can Do to Improve Your PCR Results”