If asked what are the differences between a grandfather and his newborn granddaughter, I would reply with the obvious ones: size (the grandfather is larger than his granddaughter), condition of the skin (babies have soft, smooth skin and elders have age spots and wrinkles) and life expectancy. Other visual cues may seem more similar than different. For example, grandfather and granddaughter may both lack hair on their heads or need assistance to move from one place to another. However, both baby and elder are a product of the genes expressed from their genome even if the exact sequence is not identical between them. Because genes are expressed differently over a human’s lifetime, Heyn et al. decided to examine the methylation profile in the genomes of newborns and individuals 89 years old or older. Continue reading “Methylation of Your Genome Decreases as You Age”
epigenetics
The Link Between Childhood Adversity and Cellular Aging
 Adversity and stress are known risk factors for psychiatric disorders, cardiovascular and immune disease, cognitive decline and other health problems. The long-term negative effects of adversity seem to be greatest if the traumatic events were experienced during childhood, when the brain and other biological systems are developing and maturing. Researchers are working to identify the mechanisms involved and have identified telomere shortening as one possible mechanism by which adversity increases morbidity and mortality. Continue reading “The Link Between Childhood Adversity and Cellular Aging”
Adversity and stress are known risk factors for psychiatric disorders, cardiovascular and immune disease, cognitive decline and other health problems. The long-term negative effects of adversity seem to be greatest if the traumatic events were experienced during childhood, when the brain and other biological systems are developing and maturing. Researchers are working to identify the mechanisms involved and have identified telomere shortening as one possible mechanism by which adversity increases morbidity and mortality. Continue reading “The Link Between Childhood Adversity and Cellular Aging”
Use of Cell-Free Protein Expression for Epigenetics-Related Applications
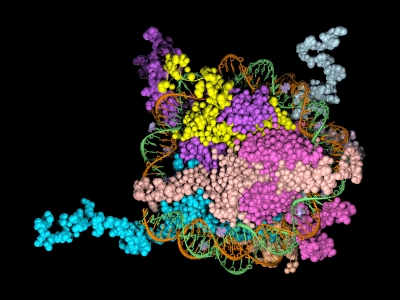 Epigenetics is the study of the processes involved in the genetic development of an organism, especially the activation and deactivation of genes. One way that genes are regulated is through the remodeling of chromatin. Chromatin is the complex of DNA and the histone proteins with which it associates. The conformation of chromatin is profoundly influenced by the post-translational modification of the histone proteins. These modifications include acetylation, methylation, ubiquitylation, phosphorylation and sumolyation. The following references illustrate the use of cell-free expression to characterize this process.
Epigenetics is the study of the processes involved in the genetic development of an organism, especially the activation and deactivation of genes. One way that genes are regulated is through the remodeling of chromatin. Chromatin is the complex of DNA and the histone proteins with which it associates. The conformation of chromatin is profoundly influenced by the post-translational modification of the histone proteins. These modifications include acetylation, methylation, ubiquitylation, phosphorylation and sumolyation. The following references illustrate the use of cell-free expression to characterize this process.
Shao, Y. et al. (2010) Nucl. Acid. Res. 38, 2813–24.
Carbonic anhydrase IX (CAIX) plays an important role in the growth and survival of tumor cells.The MORC proteins contain a CW-type zinc finger domain and are predicted to have the function of regulating transcription, but no MORC2 target genes have been identified. CAIX mRNA to be down-regulated 8-fold when MORC2 was overexpressed. Moreover, MORC2 decreased the acetylation level of histone H3 at the CAIX promoter. Among the six HDACs tested, histone deacetylase 4 (HDAC4) had a much more prominent effect on CAIX repression. Assays showed that MORC2 and HDAC4 were assembled on the same region of the CAIX promoter. Interaction between MORC2 and HDAC 4 were confirmed by using cell free expression of MORC2 and GST-HDAC (GST pull-downs). Cell-free expression was also used to express MORC2 proteins to determine through gel shifts the binding location on the CAIX promoter region (gel shift experiments)
Denis, H. et al. (2009) Mol. Cell. Biol. 29, 4982–93.
The recent identification of enzymes that antagonize or remove histone methylation offers new opportunities to appreciate histone methylation plasticity in the regulation of epigenetic pathways. PAD4 was the first enzyme shown to antagonize histone methylation. Very little is known as to how PADI4 silences gene expression. Through the use of cell-free expression to express both PAD4 and HDAC1 proteins and E. coli expression of GST fusions of PAD4 and HDAC1, pulldown experiments confirmed by in vivo experiments that PADI4 associates with the histone deacetylase 1 (HDAC1), and the corresponding activities, associate cyclically and coordinately with the pS2 promoter during repression phases.
Brackertz, M. et al. (2006) Nucl. Acid. Res. 34, 397-406.
The Mi-2/NuRD complex is a multi-subunit protein complex with enzymatic activities involving chromatin remodeling and histone deacetylation. The function of p66α and of p66β within the multiple subunits has not been addressed. GST-fused histone tails of H2A, H2B, H3 and H4 were expressed in E. coli used in an in vitro pull-down assay with radioactively labeled p66-constructs expressed using cell free systems. Deletions at the C terminus noted reduced binding of p66 where as deletions at the N terminus did not affect binding. Also observed was that acetylation of histone tails reduces the association with both p66-proteins in vitro.
Zhou, R. et al. (2009) Nucl. Acids. Res. 37, 5183–96.
Lymphoid specific helicase (Lsh) belongs to the family of SNF2/helicases. Disruption of Lsh leads to developmental growth retardation and premature aging in mice. However, the specific effect of Lsh on human cellular senescence remains unknown. In vivo results noted that Lsh requires histone deacetylase (HDAC) activity to repress p16INK4a. Moreover, overexpression of Lsh is correlated with deacetylation of histone H3 at the p16 promoter. In vitro pull-downs using cell free expression and GST fusions from E. coli were used to collaborate interactions between Lsh, histone deacetylase 1 (HDAC1) and HDAC2 observed in vivo.
Ovarian Clear Cell Carcinoma: Frequently Occuring Mutations
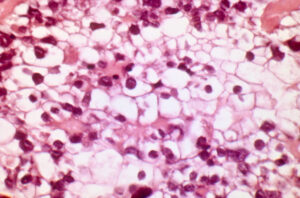
In a 2010 study of ovarian clear cell carcinoma , Jones and colleagues report a discovery of a gene that is mutated in high frequency and is involved in epigenetic regulation of gene expression.
Continue reading “Ovarian Clear Cell Carcinoma: Frequently Occuring Mutations”The Ongoing Legacy of the Human Genome Sequence

When the first draft sequence of the human genome was announced, I was a research assistant for a lab that was part of the Genome Center of Wisconsin where I created shotgun libraries of bacterial genomes for sequencing. Of course, the local news organizations were all abuzz with the news and sought opinions on what this meant for the future, including that of the lab’s PI and oddly enough, my own. While I do not recall the exact words I offered on camera, I believe they were something along the lines of this is only the first step toward the future of human genetics. Ten years later, we have not fulfilled the potential of the grandiose words used to report the first draft sequence but have gained enough knowledge of what our genome holds to only intrigue scientists even more.
Continue reading “The Ongoing Legacy of the Human Genome Sequence”
