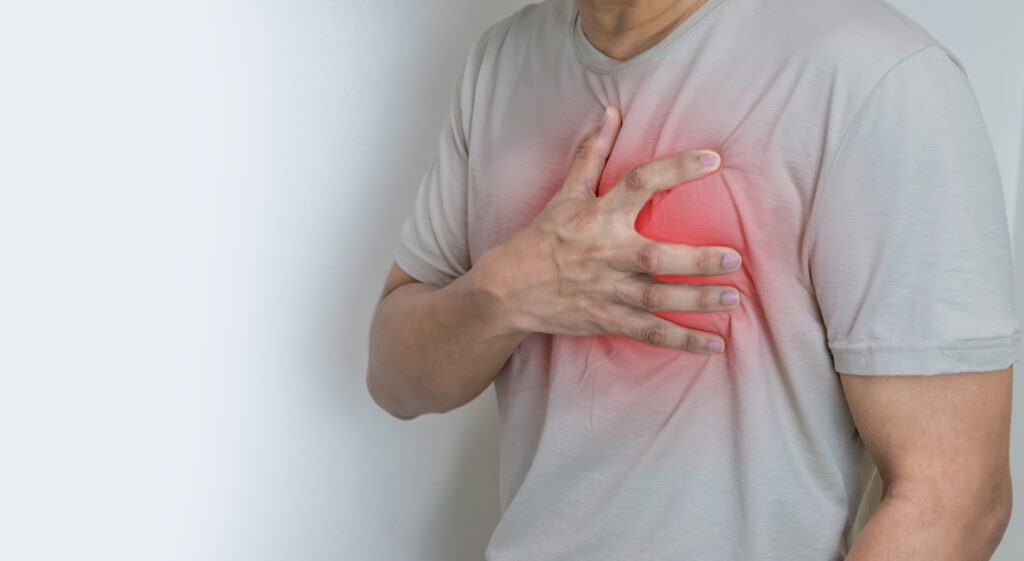
The human body has an incredible capacity for self-repair. Our skin can regenerate after a small cut, bones can heal after fractures and even the liver can regrow to its original size after 70% is lost or removed (3). However, when it comes to the heart, the story is very different. As Miley Cyrus once sang, “nothing breaks like a heart” – and science agrees. Unlike other organs, the heart has almost no ability to regenerate itself after injuries. In instances like myocardial infarctions, more commonly known as heart attacks, large amounts of cardiomyocytes (CMs)—the cells responsible for heart muscle contraction—are lost and cannot be regenerated, causing the formation of non-regenerative fibrotic scar tissue and, ultimately, decline in heart function (1).
Continue reading “Do Zebrafish Hold the Key to Heart Regeneration? “