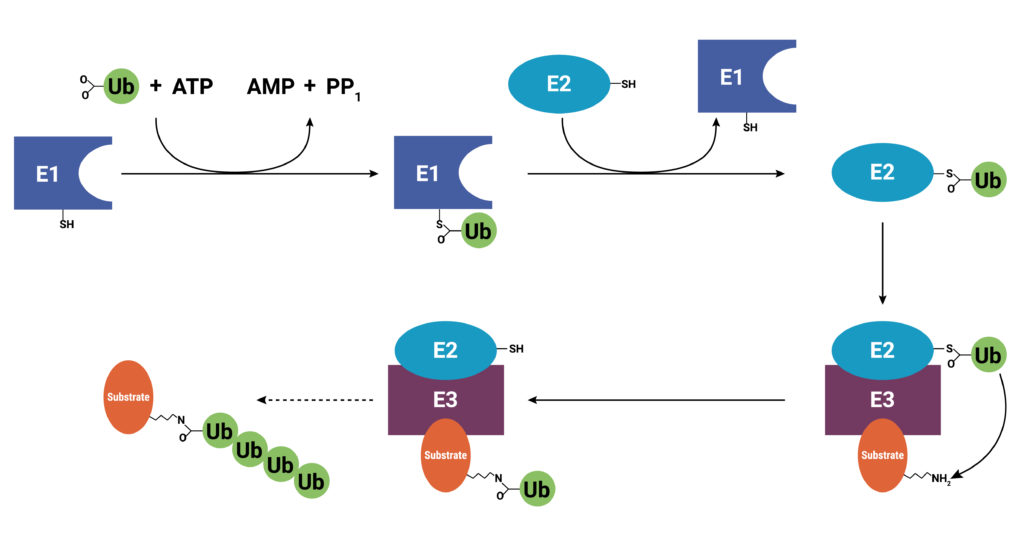
With use and time things wear out. Tires get worn on a car, and you have the old tires removed, recycled, and replaced with new ones. Sometimes a part or piece of something isn’t made properly. For instance, if you are assembling a piece of furniture and you find a screw with no threads, you throw it out and get a screw that was made properly. The same thing holds true for cells. Components wear out (like tires) or get improperly made (a screw with no threads), or they simply have a limited lifetime so that they are available in the cell only when needed. These used and worn components need to be removed from the cell. One system that allows cells to recycle components and remove old or improperly functioning proteins is the Ubiquitin-Proteasome System (UPS). The UPS system relies on a series of small peptide tags, ubiquitin, to mark a protein for degradation. Researchers are now harnessing the UPS to target aberrant proteins in diseased cells through PROteolysis TArgeting Chimeras or PROTACs. PROTACs hold promise as highly efficacious therapeutics that can be directed to eliminate only a single protein. To take full advantage of the power of PROTACs, researchers need to understand the molecular underpinnings that are responsible for successful protein degradation. Here we review a paper that seeks to develop a computer model for predicting whether PROTAC ternary complex formation leads to ubiquitination and successful degradation of a target protein.
Addressing the Intractable Target
Research to understand diseases including cancers, neurodegeneration, and auto-immune conditions has revealed that in many disease states, affected cells produce growth factors or enzymes that are constitutively active (“always on”). These proteins are targets for small molecule inhibitors that bind specific sites preventing the constitutive activity or signaling. More recently, biologics, or protein-based therapeutics, including monoclonal antibodies (mAb), have been developed that can bind and block inappropriate signaling pathways, especially those that allow cancer cells to escape immune system surveillance.
Unfortunately, up to 85% of targets have proven intractable to small molecule inhibitors, or they are not suitable for a biologics approach. Oftentimes, the target protein doesn’t have a great place to bind a small molecule, so even though inhibitors might exist they cannot bind well enough to be effective. Or, as in the case of many cancers, the diseased cell manages to overcome the effect of the inhibitor by overexpressing the target. Still other aberrant proteins associated with diseases haven’t gained function to cause a disease; they have instead, lost function, so designing an inhibitor of the protein is not a workable strategy. Enter the PROTAC.
Need some additional background about PROTACs? Read this blog to understand more of the basic molecular biology of PROTACs.
What are PROTACs, Exactly, and Why Are They Useful for Difficult Therapeutic Targets?
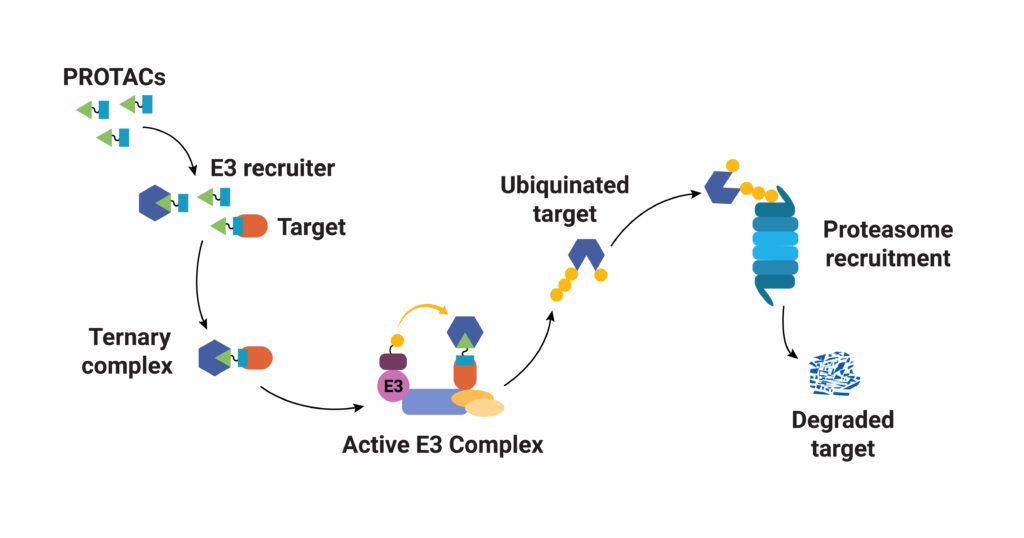
As their name implies, PROTACs are hybrid (chimeras), bifunctional molecules. One moiety (the warhead ligand) attaches to the target protein, and the other (the E3 ligand) binds to a component of the ubiquitin E3 ligase complex. The two parts of a PROTAC are connected by a linker. Ideally, a PROTAC would engage the target protein and E3 ligase component simultaneously to form a “ternary complex”. Formation of the PROTAC ternary complex results in ubiquitination of the target protein and subsequent recruitment of the proteasome for degradation. Because the PROTAC binds to the specific protein of interest and targets only that protein for degradation, using PROTACs as therapeutics provides a highly specific way to treat a disease. In theory any protein can be tagged for degradation using a cell’s existing machinery. Proteins that do not have a good binding site for an inhibitor or those that disrupt normal cellular function through a loss of function can still be specifically targeted for degradation using a PROTAC.
Most Likely to Succeed: Predicting Productive PROTAC Ternary Complexes
As promising as PROTACs are, there is much to be learned about the factors that influence successful degradation of a target protein. What are the rate-limiting steps to successful protein degradation? Can we understand how the molecular structure and conformation of the PROTAC ternary complex affect ubiquitination and, therefore, protein degradation? As we learn more about the characteristics of successful PROTACs, we will be able to create computational models to help us design more effective PROTACs for drug discovery and development.
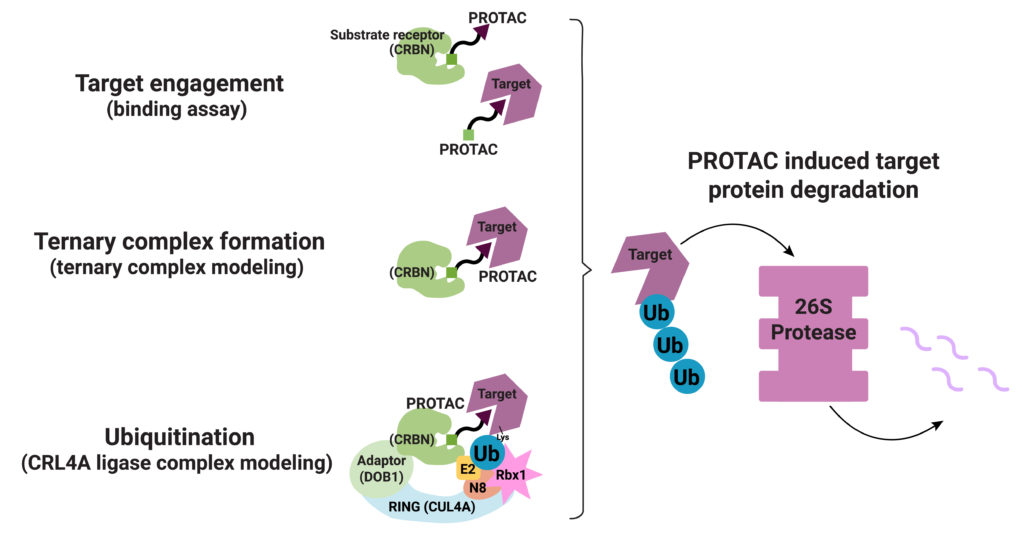
PROTAC ternary complex formation is required for successful targeted protein degradation, but ternary complex formation does not always lead to successful degradation. In fact, studies show that the conformational flexibility of the ternary complex and the availability of lysine residues in the target protein for ubiquitination are both factors that influence successful protein degradation (Crews, C. et al.). In a recent paper Bai and collaborators present work to understand successful ternary complex formation using computational models. Specifically, they set out to develop a modeling process to predict target protein ubiquitination induced by cereblon (CRBN-based) PROTACs.
Their process involved a three-step approach. First, they generated structures for the ternary complex (Target-PROTAC-CRBN) that included the many ways these three components can interact. Next, they modeled the possible CRL4A ligase conformations. They aligned each conformation from their ternary complex assemblies with each conformation of the CRL4A ligase complex ensemble. They then classified each resulting ensemble of target/PROTAC/CRBN/DDB1/ CUL4A/Rbx1/NEDD8/E2/Ub complex as “productive” or “unproductive” based on the proximity of ubiquitin to the exposed lysines on the target. Structures considered “unproductive” were excluded from further study.
Validating the Model Using Reported PROTAC Structures
Bai et al. next looked to reported CRBN-based ternary complex crystal structures to validate their model: 6BOY (Brd4/dBET6/CRBN), 5HXB (GSTP1/CC-885/CRBN) and 5FQD (CKla/lenalidomide/CRBN). They aligned each solved ternary complex structure with five conformations of their CRL4A ligase ensemble. In every case, they were able to identify at least one lysine residue on the target protein that met their criteria for proximity to the Ub in the ligase complex.
Next, the researchers looked to the CRBN-based, pan-kinase PROTAC inhibitor, TL12-186. This PROTAC induces degradation of many kinases, including many Cyclin-Dependent Kinases (CDKs). Previous work to generate degradation profiles of 16 CDKs indicated that TL12-186-induced targeted protein degradation occurred with varied efficiencies even among highly conserved members of the CDKs. This PROTAC and the diverse degradation profiles of these CDKs provided an excellent system to test their model. Bai et al. applied their computational model to generate CRBN/TL12-186/CDKx complexes, classifying them as productive or unproductive, and predicted the ubiquitination sites on the kinases. They found that the percentage of productive ternary complexes out of the total formed correlated with degradation efficiency.
Validating the Model using the NanoBRET® Ubiquitination Assay and Site-Directed Mutatgenesis
Next, Bai et al. measured ubiquitination of CDKs using a NanoBRET® Ubiquitination Assay to validate that the PROTAC ternary complexes they had identified as “productive” did result in ubiquitination of the target CDK. They used CRISPR-edited HEK293 cell lines expressing endogenous HiBiT-CDKx proteins, ectopically expressed LgBiT and a HaloTag®-Ub fusion. Upon complementation of HiBiT and LgBiT, a bright luminescent signal is produced, and this luminescent signal can serve as an energy source for the HaloTag fluor (Bioluminescent Resonance Energy Transfer, BRET) if the CDKx-HiBiT-LgBiT and HaloTaq-Ub are in proximity to each other. When the CDK target is ubiquinated, a BRET signal is produced.
They found that, when the target CDKs are treated with the TL12-186 PROTAC, the different ubiquitination profiles for each protein supported the model predictions and matched previously published results of degradation efficiency. Further, to test the model’s predictions of ubiquitinated lysine residues, the researchers performed site-directed mutagenesis was performed to substitute arginine residues for these lysine residues to see if ubiquitination or protein degradation is changed by these substitutions. For two of the three CDKs investigated, ubiquitination was either abolished or significantly reduced with mutation of a single lysine residue. The third CDK protein showed an increase in ubiquitination upon lysine mutation, which the authors posit may have resulted from an increase in affinity for other nearby lysine residues following mutation.
Summary
Ternary complex formation is necessary but not sufficient for target protein degradation. In this research, Bai et al. have addressed questions to better understand the rate-limiting steps between ternary complex formation and target protein degradation. They have developed a structure-based computer model approach to predict the efficiency and sites of target protein ubiquitination by CRNB-binding PROTACs. Such models will allow a more complete understanding of PROTAC-directed degradation and allow crafting of increasingly effective and specific PROTACs for therapeutic applications.
Another major feature of this research is that it a result of collaboration between research groups at Amgen, Inc. and Promega Corporation. In the past commercial research laboratories have shied away from collaboration, but the last several years have found researchers more open to collaborative work. This increased collaboration allows scientists to bring their different expertise to a problem or question and speed up discovery. According to Dr. Kristin Riching, Senior Research Scientist at Promega Corporation, “Targeted protein degraders have broken many of the rules that have guided traditional drug development, but it is exciting to see how the collective learnings we gain from their study can aid the advancement of this new class of molecules to the clinic as effective therapeutics.”
Literature Reviewed
Bai, N. , Riching K.M. et al. (2022) Modeling the CRLRA ligase complex to predict target protein ubiquitination induced by cereblon-recruiting PROTACs. J. Biol. Chem.
The researchers NanoBRET assays as part of their model validation. Learn more about NanoBRET technology at the Promega.com website.
Michele Arduengo
Latest posts by Michele Arduengo (see all)
- The Casual Catalyst: Science Conversations and Cafes - July 18, 2024
- Cancer Moonshot: Solving Tough Problems - May 28, 2024
- Automated Sampling and Detection of ToBRFV: An Emerging Tomato Virus - April 25, 2024

One thoughtful comment