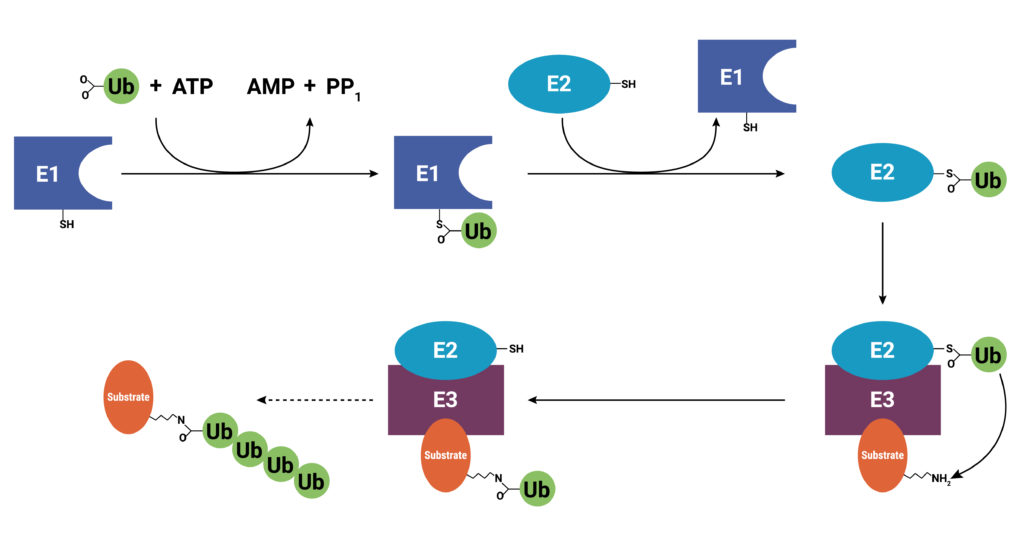
With use and time things wear out. Tires get worn on a car, and you have the old tires removed, recycled, and replaced with new ones. Sometimes a part or piece of something isn’t made properly. For instance, if you are assembling a piece of furniture and you find a screw with no threads, you throw it out and get a screw that was made properly. The same thing holds true for cells. Components wear out (like tires) or get improperly made (a screw with no threads), or they simply have a limited lifetime so that they are available in the cell only when needed. These used and worn components need to be removed from the cell. One system that allows cells to recycle components and remove old or improperly functioning proteins is the Ubiquitin-Proteasome System (UPS). The UPS system relies on a series of small peptide tags, ubiquitin, to mark a protein for degradation. Researchers are now harnessing the UPS to target aberrant proteins in diseased cells through PROteolysis TArgeting Chimeras or PROTACs. PROTACs hold promise as highly efficacious therapeutics that can be directed to eliminate only a single protein. To take full advantage of the power of PROTACs, researchers need to understand the molecular underpinnings that are responsible for successful protein degradation. Here we review a paper that seeks to develop a computer model for predicting whether PROTAC ternary complex formation leads to ubiquitination and successful degradation of a target protein.
Addressing the Intractable Target
Research to understand diseases including cancers, neurodegeneration, and auto-immune conditions has revealed that in many disease states, affected cells produce growth factors or enzymes that are constitutively active (“always on”). These proteins are targets for small molecule inhibitors that bind specific sites preventing the constitutive activity or signaling. More recently, biologics, or protein-based therapeutics, including monoclonal antibodies (mAb), have been developed that can bind and block inappropriate signaling pathways, especially those that allow cancer cells to escape immune system surveillance.
Unfortunately, up to 85% of targets have proven intractable to small molecule inhibitors, or they are not suitable for a biologics approach. Oftentimes, the target protein doesn’t have a great place to bind a small molecule, so even though inhibitors might exist they cannot bind well enough to be effective. Or, as in the case of many cancers, the diseased cell manages to overcome the effect of the inhibitor by overexpressing the target. Still other aberrant proteins associated with diseases haven’t gained function to cause a disease; they have instead, lost function, so designing an inhibitor of the protein is not a workable strategy. Enter the PROTAC.
Continue reading “Using Structural Computation Models to Predict Productive PROTAC Ternary Complexes”