The concept of cell death as a normal cell fate was articulated only three years after Schleiden and Schwann introduced the Cell Theory when, in 1874, Vogt described natural cell death as an integral part of toad development (as cited in Cotter and Curtin, 2003). Since these early observations, natural cell death has been described “anew” several times. In 1885 Flemming provided the first morphological description of a natural cell death process, which we now label “apoptosis”, a term coined by Kerr and colleagues to describe the unique morphology associated with a cell death that differs from necrosis (as cited in Kerr et al. 1972).
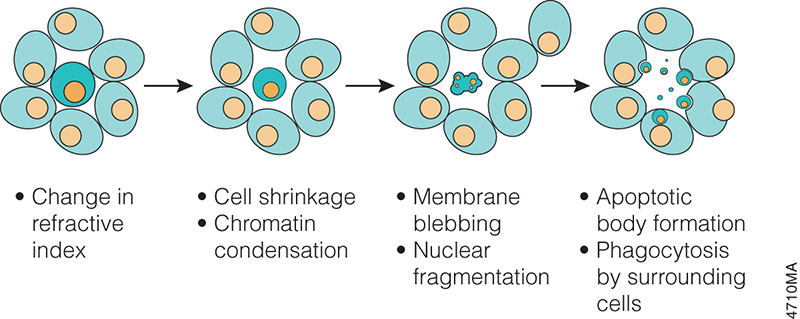
In the 1970s and 1980s, studies revealed that apoptosis not only had specific morphological characteristics but that it was also a tightly regulated process with specific biochemical characteristics. Studies of cell lineage in the nematode, Caenorhabditis elegans, showed that apoptosis was a normal feature of the nematode’s invariant developmental program (Hengartner, 1997). At the biochemical level, Wyllie showed that DNA degradation by a specific endonuclease during apoptosis resulted in a DNA ladder composed of mono- and oligonucleosomal-sized fragments (Wyllie, 1980).
These and many other studies have proven that apoptosis is a critical component of development, and when it doesn’t happen appropriately, it can be pathological, leading to cancers or other diseases. Therefore, understanding how and when apoptosis occurs and the many signals that can trigger this process is a focus of many laboratory experiments.
There are many ways to detect apoptosis in cells or tissues. This blog describes some of the most common ones.
Detecting Caspase Activity and Activation. The caspase family of cysteine proteases are the central mediators of the proteolytic cascade leading to cell death and elimination of compromised cells. Assays that directly measure caspase activity can provide valuable information for the researcher about the mechanism of death in dying cells. Luminescent cell-based assays such as the Caspase-Glo® Assays are available to detect activity of caspases involved in either the intrinsic or extrinsic apoptosis pathways. Additionally fluorescent assays are also available. Activated caspase-3 can also be detected using an in situ marker that is a fluorescent analog of the pan caspase inhibitor Z-VAD-FMK.
Detecting DNA Fragmentation. Many of the assays used to detect apoptosis analyze the characteristic DNA fragmentation that occurs during apoptosis. In apoptotic cells, the genomic DNA is cleaved to multimers of 180–200bp (based on the nucleosomal repeat length). This cleaved DNA is easily observed as a “ladder” upon analysis by gel electrophoresis. To detect this DNA fragmentation at the single-cell level, assays rely on labeling the ends of the nucleosomal fragments followed by either colorimetric or fluorescent detection. This approach, commonly called TUNEL (TdT-mediated dUTP Nick End Labeling), relies on labeling permeabilized cells to label the 3´-OH ends of the fragments with either a fluorescent or biotinylated dUTP.
Using Two or More Methods to Confirm Apoptosis. Typically, more than one method is necessary to confirm that cell death is occurring via apoptosis. Markers of apoptosis such as caspase activity may be expressed only transiently. Therefore, to determine if apoptosis is the primary mechanism of cell death, understanding the kinetics of the cell death process in your model system is critical. If detailed information on the mechanism of cell death is desired, the duration of exposure to the toxin, the concentration of the test compound and the choice of assay endpoint become critical.
For more information about detecting apoptosis, visit the Apoptosis chapter of the Promega Protocols and Applications guide.
References
- Cotter, T.G. and Curtin, J.F. (2003) Historical perspectives.In: Essays in Biochemistry. Cotter, T.G. et al. eds. 39, 1–10.
- Kerr, J.F.R., et al. (1972) Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–57.
- Hengartner, M.O. (1997) Programmed Cell Death. In: C. elegans II. Riddle, D.L. et al. eds. 383–496. (Cold Spring Harbor Laboratory Press, Plainview, N.Y.)
- Wyllie, A. H. (1980) Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 284, 555–6.
Michele Arduengo
Latest posts by Michele Arduengo (see all)
- The Casual Catalyst: Science Conversations and Cafes - July 18, 2024
- Cancer Moonshot: Solving Tough Problems - May 28, 2024
- Automated Sampling and Detection of ToBRFV: An Emerging Tomato Virus - April 25, 2024
