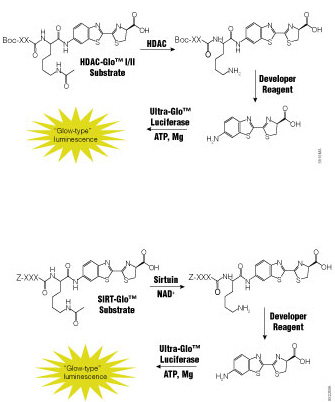
Dysfunction of histone deacetylases (HDACs) is associated with many diseases including cancers, asthma and allergies, inflammatory diseases and disorders affecting the central nervous system. Because of their involvement in such a wide range of pathologies, HDACs have become a target for drug discovery. Traditional HDAC activity assays are either isotopic or fluorescent assays using artificial substrates that are prone to artifacts or fluorescence interference. There is a need for a functional assay that is sensitive, accurate and amenable to drug-screening activities.
A recent paper by Halley et al. in the Journal of Biomolecular Screening describes the evaluation of a bioluminogenic HDAC assay, the HDAC-Glo™ I/II Assays,
The authors first used the HDAC-Glo™ I/II Assay and SIRT-Glo™ assay to determine substrate Km and IC50 values for standard inhibitors. For all enzymes assayed (HDAC-I, HDAC-3, HDAC-6 and SIRT-1), the Km values obtained using the luminogenic assays were comparable to those reported in the literature. Furthermore, the assays tolerated well the concentrations of DMSO used to deliver known HDAC inhibitors. The authors used the assays to generate dose-response curves and IC50 values for standard inhibitors of several HDAC isoforms. They indicate that all data sets were of high quality and that the IC50 values obtained were comparable to those reported in the literature.
The real test of the assays though is how they perform in high-throughput screening. Can the assays be performed in a miniaturized format? How long do the assays take, and can they be performed with minimal manipulation? Is the signal stable enough for batch-plate processing? Are data obtained from screening robust? That is: Is there minimal variability with maximal dynamic range (measured by Z´ factor)? To address these questions, the authors screened four HDAC isoforms (HDAC-1, HDAC-3, HDAC-6 and SIRT-1) in duplicate against 640 FDA-approved drugs, and they screened HDAC-6 and SIRT-1 against the Hypha Discovery MycoDiverse natural products library. For both screens, “hits” were classified as compounds that showed >50% inhibition at their screening concentration. For the 640-drug screen, the confirmation of hits was >80% and the false-positive hit rate was0.65 for the HDAC-6 screen and >0.85 for the SIRT-1 in the Hypha Discovery MycoDiverse natural products library screen. A Z´-factor of 0.5 or greater is indicative of an assay with low data variability but good dynamic range.
The authors conclude that the HDAC-Glo™ I/II and SIRT-Glo™ Assays met the criteria of sensitivity, signal stability, low background, DMSO tolerance, data variability and dynamic range, and scalability required for a suitable drug-screening assay. Additionally, the authors note that the assays required relatively low concentrations of enzymes (nM range), allowing identification of potent inhibitors and miniaturization to 10µl assay volume. The assays also yielded Km and IC50 values consistent with those in the current literature. They also note that the quality of hits obtained from the Hypha Discovery MycoDiverse natural products screening was such that purification and characterization of hits from this initial screen is ongoing.
Literature Reviewed
Halley, F. et al. (2001) A Bioluminogenic HDAC Activiyt Assay: Validation and Screening. J. Biol. Scr. 16 (10): 1227–35.
Michele Arduengo
Latest posts by Michele Arduengo (see all)
- Automated Sampling and Detection of ToBRFV: An Emerging Tomato Virus - April 25, 2024
- On-site, In-house Environmental Monitoring to Obtain Species-Level Microbial Identification - August 28, 2023
- 2023 Promega iGEM Grant Winners: Tackling Global Problems with Synthetic Biology Solutions - July 5, 2023