In targeted protein degradation (TPD), timing is everything. Understanding not just whether a degrader works—but how fast, how thoroughly and how sustainably—can dramatically influence early discovery decisions. Dr. Kristin Riching (Promega) dove into the real-time world of degradation kinetics in the webinar: Degradation in Motion: How Live-Cell Kinetics Drive Degrader Optimization, sharing how dynamic data provides a clearer view of degrader performance than traditional endpoint assays.
Whether you’re exploring your first PROTAC or optimizing a molecular glue series, the expertise offered in Dr. Riching’s presentation gives you actionable insights that will help you connect kinetic data to better therapeutic design.
Why Static Assays Fall Short
Traditional degradation assays offer a snapshot of what is happening in your cells that is useful, but often incomplete. These static measurements can miss key phases of the degradation process, including delay in onset, partial degradation or rapid protein recovery. For TPD studies, where the timing and completeness of degradation are central to therapeutic effect, static endpoints may hide meaningful distinctions between compounds.
How Live-Cell Kinetics Bring Degradation to Life
Kinetic profiles offer a lens into degrader performance that goes beyond potency, revealing timing, sustainability and cellular response potential. Continuous, live-cell monitoring using HiBiT-tagged endogenous proteins provides full degradation curves that capture:
- Degradation onset and rate
- Maximum depth of degradation (Dmax)
- Duration of protein knockdown and recovery
These metrics don’t just provide a mechanistic view—they translate directly into structure-activity relationship (SAR) decisions and compound ranking.
Turning Data into Design Decisions
Kinetic parameters provide a quantitative framework to compare compounds, even those with similar endpoint profiles. In SAR workflows, this granularity helps identify structural features that accelerate degradation or extend duration, which are both key for selecting clinical candidates.
In the webinar, Dr. Riching illustrated how kinetic data links to downstream phenotypes. For example, faster degraders often correlate with stronger, more durable biological responses, making kinetics a powerful predictor of compound efficacy.
Streamlining Degrader Workflows
One of the biggest challenges in implementing live-cell kinetic studies is balancing scientific depth with workflow simplicity. The good news? Recent innovations such as the bioluminescent HiBiT protein tagging system are lowering the barrier to entry, enabling broader adoption of kinetic profiling—even in early discovery settings.
The HiBiT system and related tools can help streamline degrader workflows across key steps:
- CRISPR/Cas9-compatible HiBiT-tagging enables rapid, precise tagging of endogenous proteins. This eliminates the need to overexpress artificial constructs, thereby preserving native cellular context and making data more physiologically relevant. HiBiT-tagged cell pools can be generated in just a few days, greatly accelerating experimental timelines.
- Pronect™ Degradation App automates the analysis of degradation curves. It calculates Dmax,degradation rate, and recovery kinetics—saving time, removing manual errors and enabling consistent comparison across compounds or targets. The platform also helps non-specialists quickly extract meaningful data, making complex kinetic analysis more accessible.
- HiBiT mRNA delivery protocols offer a reagent-ready approach to express LgBiT in nearly any cell line. This ensures fast, uniform delivery with minimal optimization, supporting reproducibility across different targets or cell models, and allowing users to easily transition between endpoint and kinetic analysis using the same HiBiT knock-in cell line depending on their stage of research
Together, these tools support a modular, end-to-end approach to degrader analysis—making it easier to generate, analyze and act on kinetic data without building custom workflows from scratch. They help you focus on insight, not infrastructure, providing valuable answers whether you’re evaluating a single degrader or screening a focused library.
See It in Action: Case Studies
Using the BET family proteins—BRD2, BRD3, and BRD4— Dr. Riching showcased how kinetic analysis reveals subtle but crucial differences. For instance, BRD2 rapidly reaccumulates after degradation, pointing to distinct feedback mechanisms. Such findings underscore the need to pair degradation kinetics with biological context for optimal compound selection.
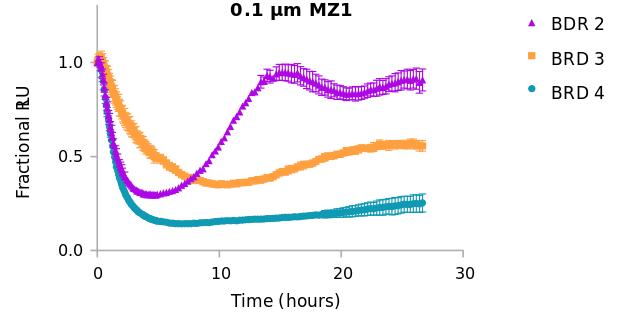
Kinetic responses of BET family members to MZ1 PROTAC. BET family members, BRD2, BRD3, and BRD4 exhibit differential degradation rate, extent and duration induced by MZ1, illustrating the need for kinetic analysis over endpoint measurements to evaluate and rank compound potency.
Ready to Add Kinetics to Your Workflow?
By answering questions about how fast, how thoroughly and howsustainably a degrader works early in the discovery process, live-cell kinetic analysis has the potential to transform how degraders are evaluated, designed and selected.
Watch the full webinar if you want to learn more about the advantages of including live-cell kinetic assays in your degrader development workflow. Gain the knowledge and tools to:
- Go beyond static assays
- Quantify degrader performance with precision
- Integrate kinetic insights into your optimization strategy
Isn’t it time to go beyond a simple yes or no and see protein degradation in motion?
Kelly Grooms
Latest posts by Kelly Grooms (see all)
- From Forever Chemicals to Ancient Proteins: Five Science Stories from 2025 - January 8, 2026
- World Wildlife Conservation Day: Reflecting on the Role of Science in Protecting Threatened or Endangered Species and Ecosystems - December 4, 2025
- Unlocking the Power of Live-Cell Kinetics in Degrader Development - September 2, 2025
