The ISOQUANT® Isoaspartate Detection Kit is intended for quantitative detection of isoaspartic acid residues in proteins and peptides, which can result from the gradual, nonenzymatic deamidation of asparagine or rearrangement of aspartic acid residues.
The ISOQUANT® Kit is designed to provide information regarding the global formation of isoaspartic acid residues at Asn and Asp sites, not at each site separately.
The deamidation of asparagine residues and rearrangement of aspartic acid residues is characterized by the formation of a succinimide intermediate that resolves to form a mixture of isoaspartic acid (typically 70–85%) and aspartic acid.
The rate and extent of isoaspartic acid formation can vary widely among proteins, depending on the amino acid sequence and size of the target protein. Deamidation of Asn residues has been observed most frequently at Asn-Gly and Asn-Ser sites within proteins.
The ISOQUANT® Isoaspartate Detection Kit uses the enzyme Protein Isoaspartyl ethyltransferase (PIMT) to specifically detect the presence of isoaspartic acid residues in a target protein. PIMT catalyzes the transfer of a methyl group from S-adenosyl-L-methionine (SAM) to isoaspartic acid. Spontaneous decomposition of this methylated intermediate results in the release of methanol and reformation of the succinimide.
References:
Wang, W. et al. (2012) Quantification and characterization of antibody deamidation by peptide mapping with mass spectrometry. Int. J. Mass. Spec. 312, 107–13.
Grappin, P. et al. (2011) New proteomic developments to analyze protein isomerization and their biological significance in plants. J. Proteomics, 74, 1475–82.
Yang, H. and Zubarev, R.A. (2010) Mass spectrometric analysis of asparagine deamidation and aspartate isomerization in polypeptides. Electrophoresis 31, 1764–71.
Sinha, S. et al. (2009) Effect of protein structure on deamidation rate in the Fc fragment of an IgG1 monoclonal antibody. Protein Sci. 18, 1573–84.
 I am fascinated by all the ways that scientists are taking sensitive techniques and using them to look into our past. For example,
I am fascinated by all the ways that scientists are taking sensitive techniques and using them to look into our past. For example, 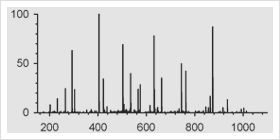
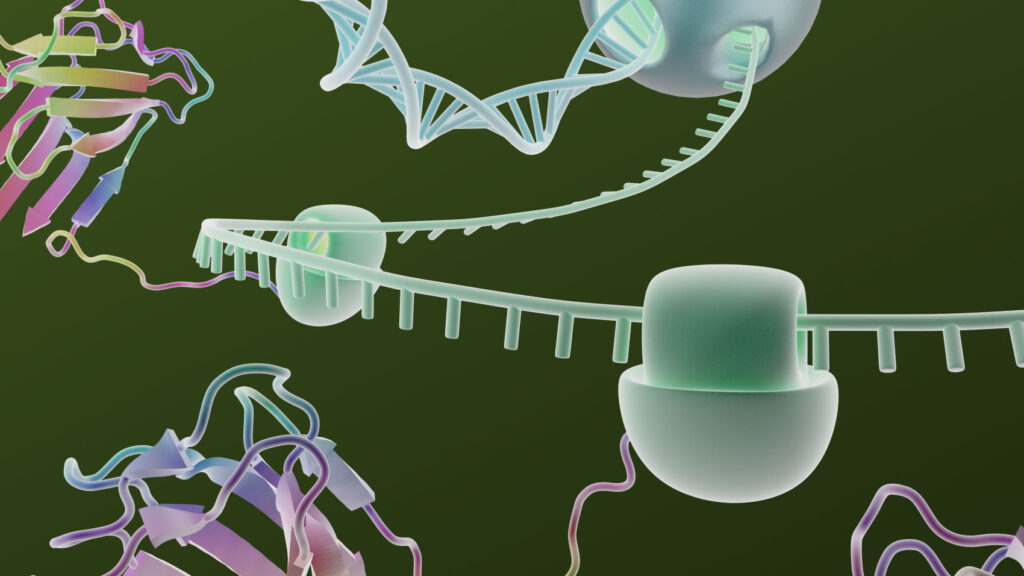
 The ability to manipulate genes and proteins and observe the effects of specific changes is a foundational aspect of molecular biology. From the first site-directed mutagenesis systems to the development of knockout mice and RNA interference, technologies for making targeted changes to specific proteins to eliminate their expression or alter their function have made tremendous contributions to scientific discovery.
The ability to manipulate genes and proteins and observe the effects of specific changes is a foundational aspect of molecular biology. From the first site-directed mutagenesis systems to the development of knockout mice and RNA interference, technologies for making targeted changes to specific proteins to eliminate their expression or alter their function have made tremendous contributions to scientific discovery.