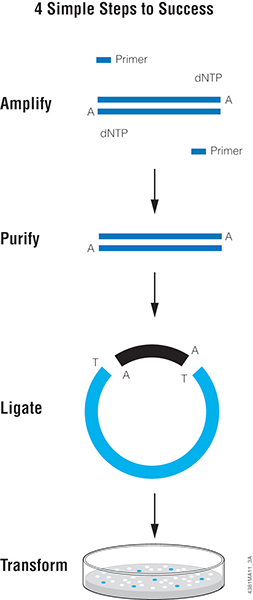
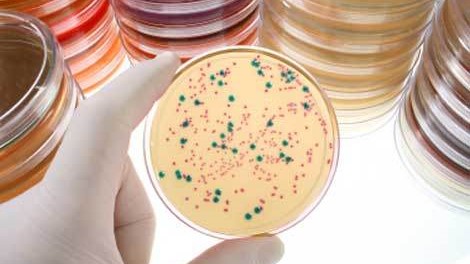
I was blasting a holiday music playlist while driving recently, and Presley’s Blue Christmas played. I couldn’t get the phrase “Christmas Cloning Blues” out of my mind, and by the time I arrived at my destination, this happened:
Cloning Blues Christmas
(to the tune of Blue Christmas by Elvis Presley)
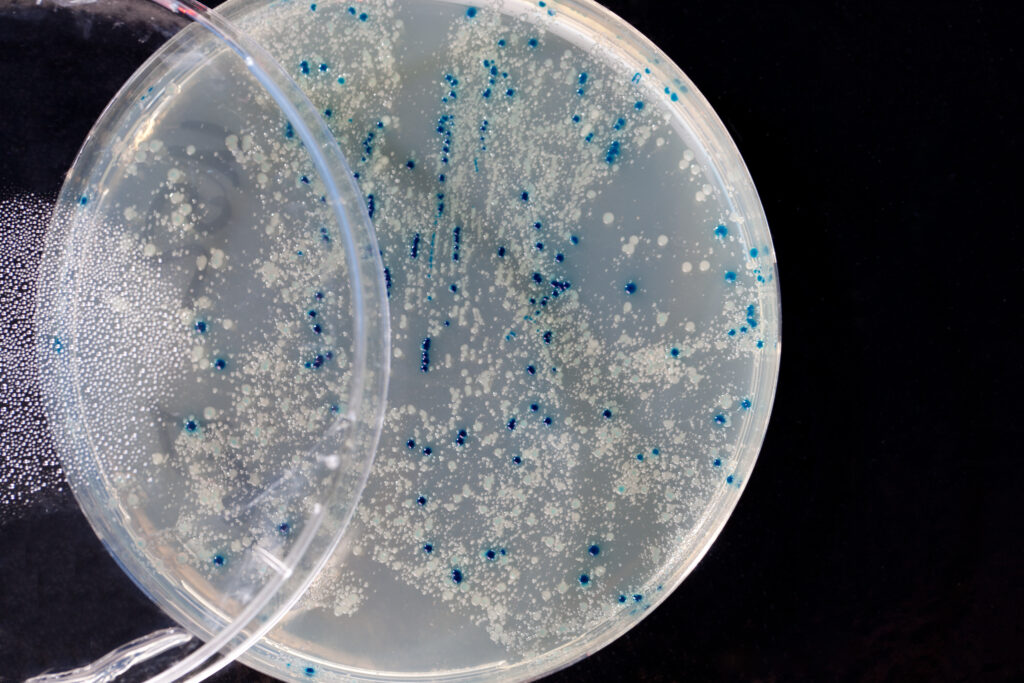
I’ll have a blue Christmas without you
Colonies so blue, insert without you
Incubating my plates at 37 degrees
Won’t be the same if you’re not in lacZ
And all those blue colonies are forming
When my lab mates’ clonings are performing
They’ll be doing alright,
With their plates all filled with white
But I’ll have a blue, blue, blue cloning
Continue reading “Avoid the Cloning Blues This Season”