In our final blog post on double-stranded RNA (dsRNA), we turn our attention to the chemical building blocks of mRNA therapeutics—modified nucleotides. These seemingly minor changes to the RNA sequence play a crucial role in the success of mRNA-based vaccines and treatments. However, they also introduce complexities in accurately detecting and quantifying unwanted dsRNA byproducts— key steps in ensuring the therapeutic efficacy of your mRNA product.
What Are Modified Nucleotides?
Modified nucleotides are ribonucleotides containing chemically altered nucleosides — like specialty ingredients swapped into a classic recipe to improve taste and nutrition. Just as a chef might use a lactose-free milk or gluten-free flour to make a dish easier to digest without changing its core structure, scientists use chemically altered nucleosides during in vitro transcription (IVT) to improve how mRNA therapies perform. These modifications replace their natural counterparts (e.g., uridine or cytidine) in the final RNA product. Their incorporation improves the performance and safety of mRNA therapeutics in several ways:
- Reduced Immunogenicity: By masking synthetic mRNA from pattern recognition receptors such as TLR3, TLR7, RIG-I, and MDA5, modified nucleotides lower the risk of innate immune activation and inflammation.
- Improved Stability: These modifications support favorable RNA folding and help protect the molecule from degradation, extending its intracellular half-life.
- Enhanced Protein Expression: Reduced immune recognition and improved RNA stability contribute to more efficient translation and higher protein output from the same dose.
- Reduced dsRNA Formation: Some modified nucleotides suppress formation of unintended dsRNA species during IVT, limiting immune triggers and improving product consistency.
Key Modified Nucleosides Used in mRNA Therapeutics
Several chemically modified nucleosides are commonly used in IVT to generate functional therapeutic mRNA:
- Pseudouridine (Ψ): An isomer of uridine, Ψ alters the glycosidic bond to improve immune evasion and reduce interferon responses.
- N1-Methylpseudouridine (m1Ψ): A methylated variant of Ψ, m1Ψ is widely used in mRNA vaccines and enhances translation by further decreasing immune recognition.
- 5-Methoxyuridine (5moU): Featuring a methoxy group on the uracil ring, 5moU reduces both antiviral and proinflammatory signaling, particularly in immune cells.
- 5-Methylcytidine (m5C): A methylated form of cytidine, m5C helps stabilize RNA structure and boost translational efficiency.
Once phosphorylated, these modified nucleosides become nucleotide triphosphates (NTPs) for incorporation during IVT.
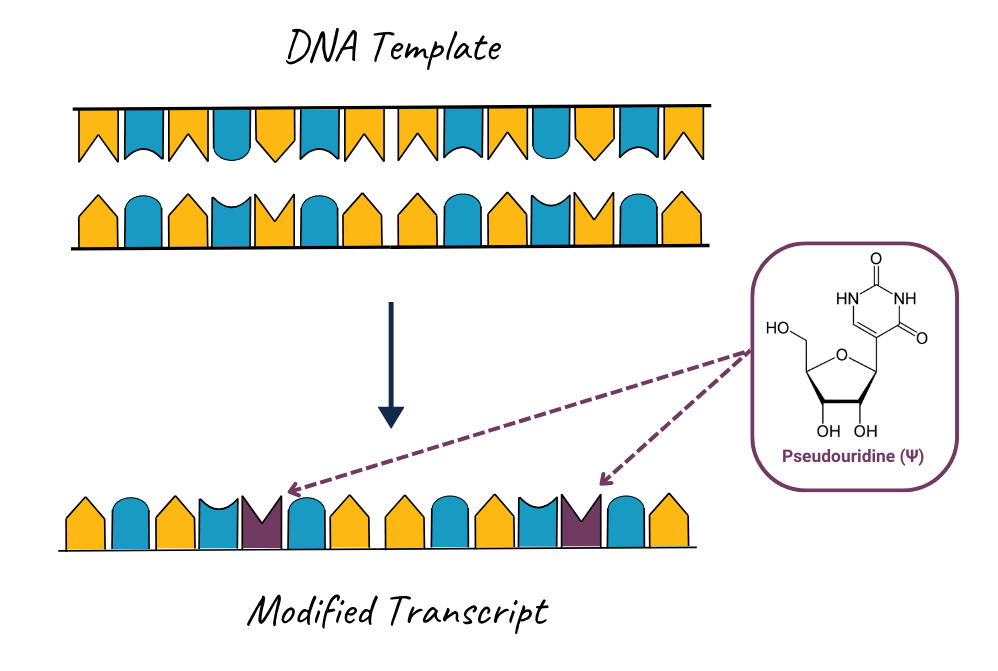
Challenges in dsRNA Detection
While these modifications enhance mRNA therapeutics, they complicate accurate quantification of dsRNA—an important impurity in IVT workflows. Traditional detection methods, such as ELISA or HTRF-based assays, rely on antibodies (e.g., J2 or K1) that recognize specific RNA structures. But RNA secondary structure and sequence—both influenced by modification—can alter antibody binding. This can result in inaccurate measurements of dsRNA, potentially leading to inappropriate dosing or missed safety concerns.
Accurately quantifying dsRNA is critical, especially since acceptable thresholds vary by application. mRNA vaccines may tolerate higher dsRNA levels to support immunogenicity, while protein replacement therapies require tighter limits to avoid off-target immune responses.
A More Sensitive and Specific Approach
To address these challenges, Promega developed the Lumit® dsRNA Detection Assay, a luminescence-based, antibody-free solution that quantifies dsRNA with high sensitivity and specificity. Using a proprietary dsRNA-binding domain linked to NanoBiT® luciferase, this assay avoids the sequence and structure biases seen with antibody-based methods.
Importantly, the Lumit assay has been demonstrated for use with dsRNA standards containing common nucleotide modifications such as Ψ, m1Ψ, and 5moU. This enables accurate quantification in modified mRNA samples. For best results, use dsRNA standards that match the nucleotide modifications present in the mRNA therapeutic or vaccine you’re analyzing. Doing so helps you ensure that assay response reflects the true content of dsRNA in the final product.
Modified nucleotides are critical tools for improving mRNA therapeutic performance. But they also demand more thoughtful analytical strategies. The Lumit® dsRNA Detection Assay offers a robust, accurate, and modification-tolerant solution—enabling developers to manage dsRNA with greater confidence across diverse therapeutic applications.
Complete the dsRNA Blog Series
This wraps up our three-part series on dsRNA in mRNA therapeutics. If you missed these earlier blogs, start by exploring the biological role of dsRNA and why it matters for mRNA therapeutics in Blog #1. Then, learn how dsRNA contaminants can arise during in vitro transcription (IVT) and strategies to minimize them in Blog #2.

Looking for a more reliable way to detect dsRNA in your RNA production? The Lumit® dsRNA Detection Assay offers a sensitive, antibody-independent solution to help you confidently measure dsRNA byproducts in mRNA therapeutic development. Learn more about how streamlined detection can support better QC decisions at every stage.
References
- Liu A and Wang X (2022) The Pivotal Role of Chemical Modifications in mRNA Therapeutics. Front. Cell Dev. Biol. 10:901510. doi: 10.3389/fcell.2022.901510
- Kariko, K., Muramatsu, H., Ludwig, J., and Weissman, D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Research, 2011, Vol. 39, No. 21
- Kellie D. Nance and Jordan L. Meier. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Central Science 2021 7 (5), 748-756
- Moradian, H., Roch, T., Anthofer, L., Lendlein A., Gossen, M. Chemical modification of uridine modulates mRNA-mediated proinflammatory and antiviral response in primary human macrophages. Molecular Therapy – Nucleic Acids. 2022. Vol 27. 854-869
