This is the second in a series of blog posts covering topics to consider when designing and performing cell-based assays. In the first installment, we discussed the importance of choosing the right cell type for your assay. Here we will discuss how cell culture conditions affect cell-based assays.
Consistent, careful handling of cells is crucial for reliable, repeatable results.
Your cell culture medium is perhaps the most basic component of cell culture. The medium you use should be fresh and consistent (from the same manufacturer). Because serum supplements are complex biological solutions, they vary from vendor to vendor and even from lot to lot from the same vendor. Be sure to note the lot and source of all serum supplements and the medium used to culture the cells used for any experiments. This will help you replicate experiments later and may be important if you need to troubleshoot in the future.
Be careful when you handle your cells; avoid rough pipetting. When you are pipetting cells, make sure that cells grown in suspension remain in suspension. When you are trypsinizing adherent cells, don’t over trypsinize: you can kill your cells. However, if your trypsinization isn’t effective, you will end up with clumps of cells. One good piece of advice is to find someone who has experience working with the cell type you are using. Spend some time with that person to learn some of the “art” culturing those particular cells successfully. When you are counting cells, consider using trypan blue exclusion dye in addition to the hemocytometer so that you can plate the correct number of viable cells.
Contamination of cell culture by microorganisms and by other cell lines is a major concern in the cell culture lab. To avoid contamination of cultures by microorganisms such as mycoplasma, excellent sterile technique is absolutely essential. If you have never maintained cell cultures before, find the technician with the best reputation around for clean cell culture, and learn from that person.
Cell line identity has been identified as a problem in many research publications, with as many as 15–20% published studies having used cells that were misidentified or cross-contaminated with another cell line (1). Most cell line repositories, many journals and grant organizations now require genetic evidence that cells used in a study are what the authors claim. Such cell line authentication is generally accomplished using STR profiling (1,2).
As the number of cell line passages increase, the cells may change character, particularly if the cells reach confluence and you begin seeing contact inhibition signaling. Your assay results may reflect this change. Percent confluence is an important parameter when transfecting cells as well, and you will need to consider cell density and confluence if transfection is part of your experimental procedure. One suggestion is to create a bulk stock at a low passage number and then thaw a new vial for each experiment so that passage number is the same for each experiment.
Cell density can affect assay results as well. You need to consider that your seeding density and the density at the time of the experiment may not be the same. For instance, will you distribute the cells into the assay plate wells and begin the assay immediately, or will the cells be cultured for any length of time before the treatment or assay?
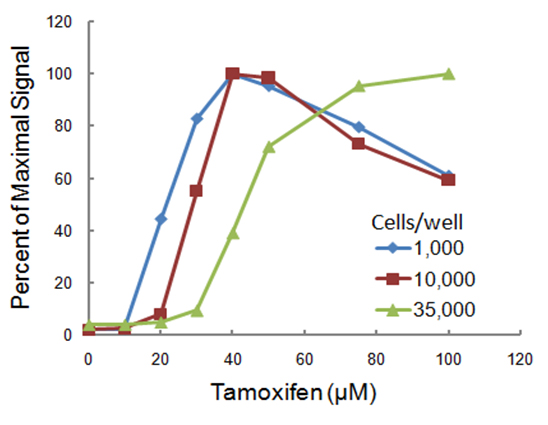
Cell density can affect dose response curves. Here we performed three dose response curves to tamoxifen in HepG2 cells seeded at three different densities. HepG2 cells were seeded at 1,000, 10,000 or 35,000 cells per well in 96-well plates and exposed to various concentrations of tamoxifen for 4 hours. Caspase-3/7 activity was measured using the luminescent Caspase-Glo® 3/7 Assay, and the results were expressed as percentages of maximum signal for each cell density. All data points represent the mean ± SD (n = 3). Reprinted with permission from Riss, T. and Moravec, R. (2004) ASSAY Drug Dev. Tech. 2, 51.
Notice that the cells seeded at the lower density were more sensitive; denser cultures may confer a “protective” effect. Because the peak activity here was seen later with lower tamoxifen concentrations, a longer exposure is necessary to see an effect when using higher density cultures. Also, overgrown cultures are going to have a higher background because of cell death and toxicity associated with aging cultures that may not be related to effects of treatment compounds.
Cell-based assays also have an optimal range of number of cells over which they will produce linear results. So when you are setting up a new experimental system, a little bit of preliminary work goes a long way to ensuring reliable results from your assay. Performing a control assay with a serial dilution of cells will help you determine the sensitivity and range of the assay. It will also provide guidance for decisions about seeding density, and timing between plating, treatment and assay. For multiplexed assays or live/dead assays, you will want to perform two control assays in parallel, one with treated cells and one with untreated cells.
These are a few of the considerations for cell culture and plating conditions that can influence cell-based assay outcomes. In the next installment, we’ll talk about treatment parameters.
Literature Cited
- Dunham, J.H. and Guthmiller, P. (2008) Doing good science: authenticating cell line identity. Cell Notes 22, 15-17.
- Oostdik, K., Petterson, A., Schagat, T. and Storts, D. Stem Cell Line Authentication and Contamination Detection. [Internet] 2009. [cited: 2012, 01, 06]. Available from:http://www.promega.com/resources/articles/pubhub/enotes/stem-cell-line-authentication-and-contamination-detection/
Resources
Cell Line Authentication Resources
Michele Arduengo
Latest posts by Michele Arduengo (see all)
- IL-6/STAT3-Regulated Long Non-Coding RNA Is Involved in Colorectal Cancer Progression - July 1, 2025
- Using Dual-Luciferase Assays to Identify the Role of Non-Coding RNAs in Disease - May 2, 2025
- An Unexpected Role for RNA Methylation in Mitosis Leads to New Understanding of Neurodevelopmental Disorders - March 27, 2025