Clostridium difficile is a bacterium that infects around half a million people per year in the United States. The infection causes symptoms ranging from diarrhea to severe colitis, and it’s one of the most common infections contracted while staying in the hospital. As the global incidence of C. diff infection has risen over the past decade, so has the pressure to develop novel therapeutic strategies. This requires a thorough exploration of all aspects of C. difficile biology.
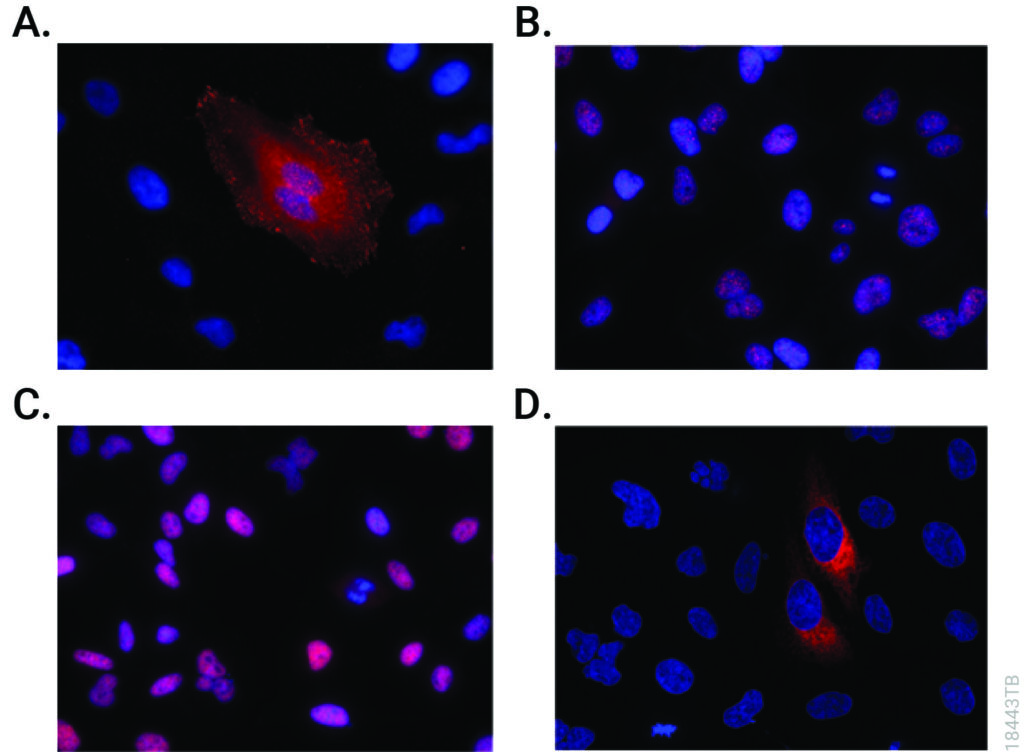
Two recent papers published by researchers at the University of Leiden have shed light on C. difficile physiology using HiBiT protein tagging. The HiBiT system allows detection of proteins in live cells using an 11 amino acid tag. The HiBiT tag binds to the complementary LgBiT polypeptide to reconstitute the luminescent NanoBiT® enzyme. The resulting luminescence is proportional to the amount of HiBiT-tagged protein that is present.
Continue reading “A Closer Look at C. difficile Biology with Luminescent Tagging”