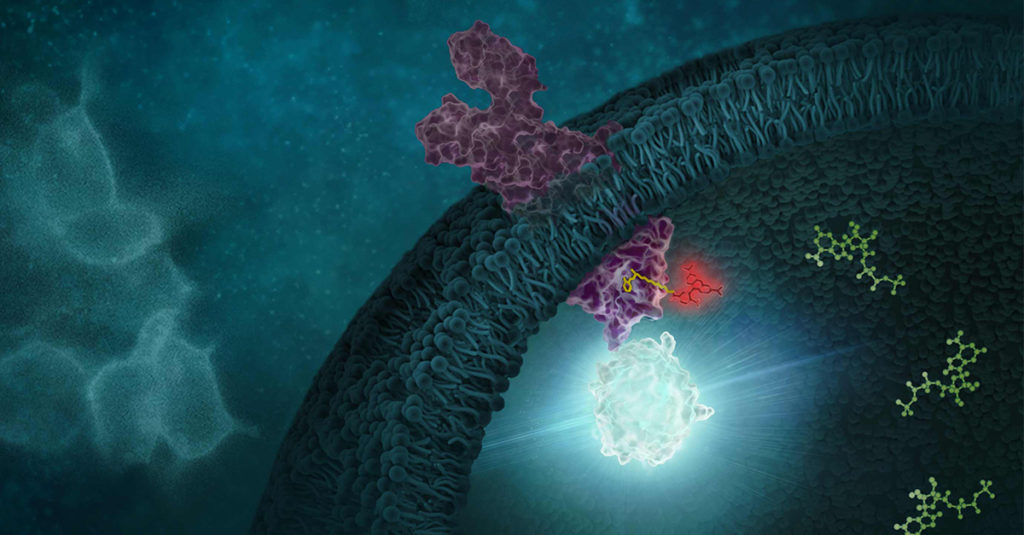
Monitoring and quantifying drug-target binding in a live-cell setting is important to bridging the gap between in vitro assay results and the phenotypic outcome, and therefore represents a crucial step in target validation and drug development (1). The NanoBRET™ Target Engagement (TE) assay is a biophysical technique that enables quantitative assessment of small molecule-target protein binding in live cells. This live-cell target engagement assay uses the bioluminescence resonance energy transfer (BRET) from a NanoLuc® luciferase-tagged target protein and a cell-permeable fluorescent tracer that reversibly binds the target protein of interest. In the presence of unlabeled test compound that engages the target protein, the tracer is displaced, and a loss of BRET signal is observed. Due to the tight distance constraints for BRET, the signal measured is specific to the target fused to NanoLuc® luciferase.
Promega offers over 400 ready-to-use assays for multiple target classes, including kinases, E3 ligases, RAS, and many others. For targets that do not have an existing NanoBRET™ TE assay, Promega offers NanoBRET™ dyes, NanoLuc® cloning vectors, and NanoBRET™ detection reagents to develop novel NanoBRET™ TE assays.
To learn more about the NanoBRET™ TE platform, see the NanoBRET™ Target Engagement Technology Page on our website.
One critical component in the development of novel NanoBRET™ TE assay is the creation of the cell-permeable fluorescent tracers (NanoBRET™ tracers) against the target protein of interest. The tracers are bifunctional, consisting of a NanoBRET™-compatible fluorophore and a target-binding moiety connected by a linker. While the NanoBRET™ 590 dyes have demonstrated consistently robust cell permeability and optimal spectral overlap with NanoLuc® for BRET, a ligand capable of binding to the target protein of interest needs to be identified to generate a NanoBRET™ tracer.
What Are DNA-Encoded Libraries?
DNA-Encoded Libraries, (DELs), have emerged as powerful tools for discovering small molecule ligands to target proteins of interest at an unprecedented scale. . owing to the ability of a DEL to enable the synthesis of larger libraries of compounds and to target proteins without any prior structural knowledge of the proteins or their ligands (2). Because each member of a DEL contains a DNA barcode and a small molecule separated by a linker, DEL is primed for discovering leads within therapeutic modalities that rely on bifunctional chemistry, such as proteolysis targeting chimeras (PROTACs). Since NanoBRET™ tracers are also bifunctional, ligands identified from DEL selections could serve as ideal candidates for developing novel NanoBRET™ tracers that can enable NanoBRET™ TE assays for new targets.
Continue reading “From Hit to Live-Cell Target Engagement Assay: DNA-Encoded Library Screens and NanoBRET™ Dye”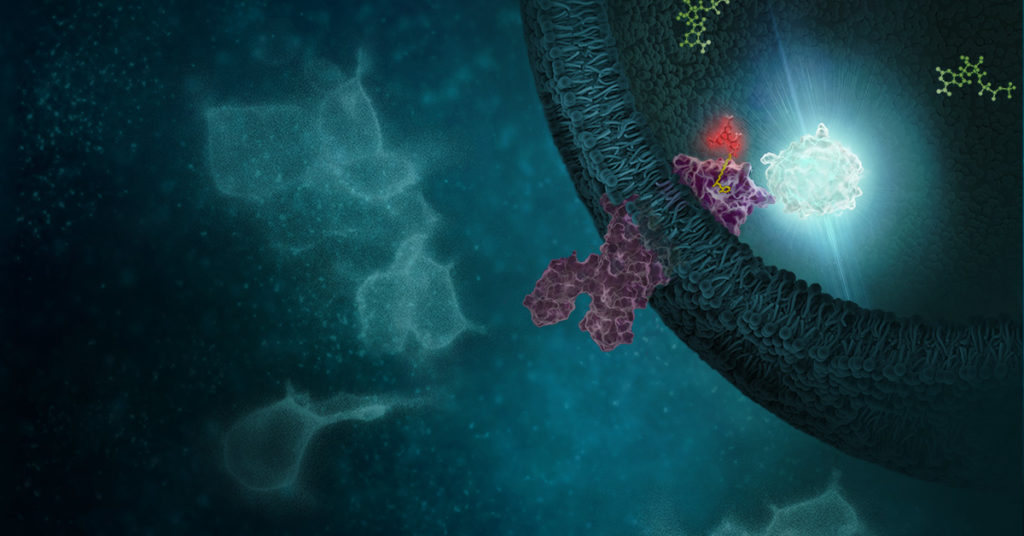

![Fig 4. Four point MMOA screen for tideglusib and GW8510. Time dependent inhibition was evaluated by preincubation of TbGSK3β with 60 nM tideglusib and 6 nM GW-8510 with 10μM and 100μM ATP. (A). Tideglusib [60 nM] in 10μM ATP. (B). GW8510 [60 nM] in 10μM ATP. (C.) Tideglusib [60 nM] at 100μM ATP. (D.) GW8510 [60 nM] at 100μM ATP. All reactions preincubated or not preincubated with TbGSK3β for 30 min at room temperature. Experiments run with 10μM GSM peptide, 10μM ATP, and buffer. Minute preincubation (30 min) was preincubated with inhibitor, TbGSK3β, GSM peptide, and buffer. ATP was mixed to initiate reaction. No preincubation contained inhibitor, GSM peptide, ATP, and buffer. The reaction was initiated with TbGSK3β. Reactions were run at room temperature for 5 min and stopped at 80°C. ADP formed was measured by ADP-Glo kit. Values are mean +/- standard error. N = 3 for each experiment and experiments were run in duplicates. Control reactions contained DMSO and background was determined using a zero time incubation and subtracted from all reactions. Black = 30 min preincubation Grey = No preincubation.](https://www.promegaconnections.com/wp-content/uploads/2016/04/journal.pntd_.0004506.g004-243x300.png)