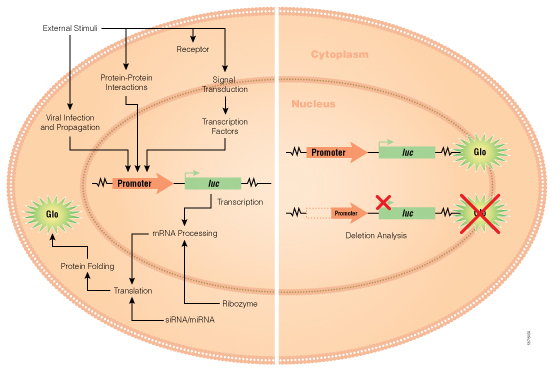
As in any biological experiment, the controls are as important, if not more, than the actual samples. There are multiple options, and researcher needs to choose the controls depending on the question they would like to ask.
If you need a yes or a no answer to the question—is this DNA fragment capable of promoter activity—then cloning a promoter like sequence upstream of GFP like protein would provide qualitative information about the activity of the promoter. This would provide a quick way to check for promoter activity. You would simply look at the cells under a fluorescent microscope to get your answer. Similarly, expression from ß-gal reporter can be determined by staining with X-gal substrate followed by microscopy. The controls for these experiments would include a) untransfected cells control and b) empty vector. These will provide information about any signal contributed by cells or media and background or leaky reporter gene expression respectively. While these approaches provide a yes or no answer about the ability of the inserted DNA element to induce gene expression, these reporters cannot be used to determine the efficacy or comparative efficacy of more than one promoter element.
Comparison of two promoter-like elements:
Let us say you need to compare the relative efficiencies of promoter A and promoter B element cloned into a luciferase reporter vector. It is important to note that these elements are cloned into two separate plasmids and each transfection experiment, however well controlled it may be, involves different plasmids from different plasmid preparations. While similar protocols may have been employed for purifying the plasmids, variables such as concentration, purity, carryover components in the sample may lead to variable transfection efficiency.
Therefore, a higher signal from promoter A than promoter B may mean that a) A is indeed a stronger promoter than B or b) the transfection efficiency of B was less than A. In order to distinguish between the two scenarios, another reporter plasmid (preferably with the same vector backbone) is co-transfected along with promoter A or promoter B containing vectors. If the signal from the co-transfected plasmid is comparable when transfected with either of the two reporter plasmids, and A > B, we can conclude that promoter A is stronger than promoter B. This technique is called normalization.
How strong do we want the normalization vector to be?
It is important to note that the function of co-transfected vector is simply that of normalization for transfection efficiency. High expression of the normalization vector is unnecessary, it is worthwhile to note that plasmid also utilizes the same cellular machinery for transcription and translation for gene expression and we certainly do not want to get a lower expression of the primary reporter activity merely because the normalization vector expression swamped the cellular resources. This is the primary reason we do not recommend a strong promoter such as CMV for normalization vector. Should you need to use a strong promoter, it is recommended that a low ratio (1:50 or lower) of primary vector: normalization vector be tested.
Small molecule screening
In most cases, the small molecule library screening is performed in a stable cell line containing a promoter or responsive element cloned upstream of a reporter gene. Stable cell lines do not require a transfection control since each cell in the clonal population contains a copy of the inserted gene in the chromosomal genome. In these cases, the most important factor to be considered while screening a library of yet-to-be characterized compounds is the effect on cellular viability. A decrease in reporter signal might arise from an actual decrease in promoter activity or simply because the compound in question caused a significant population of cells to die. Normalization of signal to the number of viable cells provides a method to interpret the results accurately. Gene expression can be normalized to viable cell number, DNA content, protein content.
Controls include untreated, vehicle-treated and positive control-treated replicates. While the difference in the signal between the untreated and the vehicle-treated provide information about any nonspecific effects such as viability or a nonspecific change in overall gene expression in the cell, the positive control usually provides valuable information about the expression capability of the reporter in the cell line, highlighting the inherent differences in cell line characteristics.
Use of experimental controls and normalization controls is essential to understanding promoter activity. In biological systems, it is equally if not more important to prove what isn’t the possibility than what is.
Reference:
Trista Schagat, Aileen Paguio and Kevin Kopish. Cell Notes 17, 9–12 (2007)Normalizing Genetic Reporter Assays Approaches and Considerations for Increasing Consistency and Statistical Significance.
Anupama Gopalakrishnan
Latest posts by Anupama Gopalakrishnan (see all)
- Mitochondrial DNA Typing in Forensics - February 25, 2015
- PowerQuant System: Tool for informed casework sample processing decisions - July 21, 2014
- Biology of Overeating and the Weight-Gain Cycle - April 28, 2014
