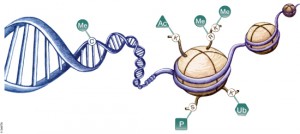
DNA is organized by protein:DNA complexes called nucleosomes in eukaryotes. Nucleosomes are composed of 147 base pairs of DNA wrapped around a histone octamer containing two copies of each core histone protein. Histone proteins play significant roles in many nuclear processes including transcription, DNA damage repair and heterochromatin formation. Histone proteins are extensively and dynamically post-translationally modified, and these post-translational modifications (PTMs) are thought to comprise a specific combinatorial PTM profile of a histone that dictates its specific function. Abnormal regulations of PTM may lead to developmental disorders and disease development such as cancer.
Continue reading “Mass Spec Analysis of PTMs Using Minimal Sample Material”ptms
Glycosyltransferases: What’s New in GT Assays?
In his 2014 blog, “Why We Care About Glycosyltransferases” Michael Curtin, Promega Global Product Manager for Cell Signaling, wrote:
“Glycobiology is the study of carbohydrates and their role in biology. Glycans, defined as ‘compounds consisting of a large number of monosaccharides linked glycosidically’ are present in all living cells; They coat cell membranes and are integral components of cell walls. They play diverse roles, including critical functions in cell signaling, molecular recognition, immunity and inflammation. They are the cell-surface molecules that define the ABO blood groups and must be taken into consideration to ensure successful blood transfusions.
The process by which a sugar moiety is attached to a biological compound is referred to as glycosylation. Protein glycosylation is a form of post-translational modification, which is important for many biological processes and often serves as an analog switch that modulates protein activity. The class of enzymes responsible for transferring the sugar moiety onto proteins is called a glycosyltransferase (GT).”
Continue reading “Glycosyltransferases: What’s New in GT Assays?”