Monoclonal antibodies (mAbs) have been widely used to eliminate undesired cells via various mechanisms, including antibody-dependent cell-mediated cytotoxicity (ADCC), complement-dependent cytotoxicity (CDC) and programmed cell death (PCD). Unlike the Fc-dependent mechanism of ADCC and CDC, certain antibody–antigen interactions can evoke direct PCD via apoptosis or oncosis. Previously, researchers have reported the specific killing of undifferentiated human embryonic stem cells (hESC) by mAb84 (IgM) via oncosis (1)
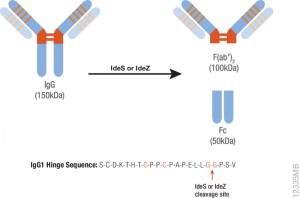
In a recent publication (2), a monoclonal antibody (mAb), TAG-A1 (A1), was generated to selectively kill residual undifferentiated human embryonic stem cells (hESC). One of the many experimental tools used to characterize the mechanism of oncosis was the fragmention of the A1 antibody with IdeS and papain.
Learn more about IdeS and IdeZ Protease available from Promega.
Papain digestion of IgG produces Fab fragments in the presence of reducing agent. F(ab)2 fragments of A1 were produced using IdeS Protease.
The results indicate that both Fab_A1 and F(ab)2_A1 bind to hESC but only F(ab)2_A1 retained hESC killing. Hence bivalency, but not Fc-domain, is essential for A1 killing on hESC.
- Choo, A.B. et al. (2008) Selection against undifferentiated human embryonic stem cells by a cytotoxic antibody recognizing podocalyxin-like protein-1. Stem Cells 26, 1454.
- Zheng, J.Y. et al. (2017) Excess reactive oxygen species production mediates monoclonal antibody-induced human embryonic stem cell death via oncosis. Cell Death and Differentiation 24, 546–58.
Are you looking for proteases to use in your research?
Explore our portfolio of proteases today.