Sometimes, when using trypsin to study a protein sequence or protein modifications, sequence coverage just isn’t quite as complete as you’d like. Looking for a protease with novel cleavage specificity or a protease that functions well in a low pH environment? Promega has a protease for that.
ProAlanase is a new site-specific endoprotease that preferentially cleaves proteins on the C-terminal side of proline and alanine amino acids. The unique cleavage specificity of ProAlanase (also known as An-PEP or EndoPro; 1–3) can help to uncover parts of the proteome not previously accessible with proteases typically used in proteomic studies.
While trypsin is the undisputed workhorse protease used in proteomics research, protein sequence information provided by tryptic digests is often incomplete. To obtain complete sequence coverage or identify missing post-translational modifications, alternative proteases are needed. Like trypsin, commonly used alternative proteases (Lys-C, Asp-N, Glu-C & Arg-C) perform cleavage at charged residues, introducing bias to regions within proteins that are digested. There is a need for site-specific proteases that cleave at unique, non-charged sites in the proteome. ProAlanase achieves this goal, preferentially cleaving proteins on the C-terminal side of proline and, to a lesser extent, alanine residues. As seen in the figure below, C-terminal cleavage at proline and alanine residues by ProAlanase accounts for up to 80-85% of the total cleavage events under optimized conditions.
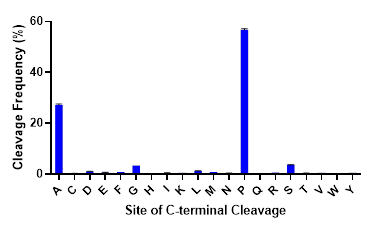
C-terminal cleavage specificity of ProAlanase. Human K562 extract was digested with ProAlanase at pH 1.5 for 2 hours at 37°C using a 1:100 enzyme:substrate ratio. Digestion was terminated by C18 cleanup. Peptides were analyzed by LC-MS/MS on a Q-Exactive Plus. Data were searched using Byonic™ software with no enzyme specified. Cleavage occurs predominantly on the C-terminal side of proline and alanine.
Are you looking for proteases to use in your research?
Explore our portfolio of proteases today.
Applications of ProAlanase
Professor Jesper Olsen’s lab group recently demonstrated that ProAlanase has proven useful in a variety of applications including phosphorylation profiling, de novo sequencing, disulfide bond mapping and paleoproteomics (See the published data here). In the case of paleoproteomics, the researchers found that digestion of Pleistocene mammoth bone samples with ProAlanase improved the protein identification rate compared to trypsin. Analysis of bone proteins is complicated by the high abundance of collagen, which is proline rich. ProAlanase digestion effectively degrades collagen into very small fragments, allowing for more efficient identification of other non-collagen proteins in the samples.
For disulfide bond mapping, they digested NISTmAb with ProAlanase under low-pH conditions which are known to minimize disulfide bond scrambling and subsequently identified numerous disulfide-bonds. Interestingly, it appears that the very acidic digestion conditions promote efficient digestion even in the absence of chemical denaturants such as urea or guanidine HCl. Monoclonal antibodies can be notoriously difficult to digest under non-reducing conditions, particularly in the absence of prior chemically-induced denaturing. This low-pH workflow for analysis of disulfide bonds with ProAlanase could be useful for the in-depth characterization of monoclonal antibodies and other biotherapeutic proteins.
Features of ProAlanase, Mass Spec Grade:
- Unique cleavage specificity: C-terminal to proline and alanine
- Fast digestion times typically 1-2 hours
- Digestion at acidic pH reduces sample prep artifacts common at basic pH
- Digestion at pH optimum (1.5) denatures many proteins, minimizing the need for additional denaturants.
Exclusively from Promega, ProAlanase is now available in low and high concentration forms:
- ProAlanase: 5 µg at 0.2 µg/µL
- ProAlanase Plus: 15 µg at 0.5 µg/µL
References
- Sebela, M. et al. (2009) Identification of N-glycosylation in prolyl endoprotease from Aspergillus niger and evaluation of the enzyme for its possible application in proteomics. J. Mass. Spectrom. 44, 1587–1595.
- Tsiatsiani, L. et al. (2017) Aspergillus niger prolyl endoprotease for hydrogen-deuterium exchange mass spectrometry and protein structural studies. Anal. Chem. 89, 7966-7973.
- van der Laarse, S. A. M. et al. (2020) Targeting proline in (phospho)proteomics. FEBS J. 287, 2979–2997.
Related Posts
Latest posts by Leah Cronan (see all)
- Mass Spec for Glycosylation Analysis of SARS-CoV-2 Proteins Implicated in Host-Cell Entry - November 10, 2020
- Proteomics from a Different Point of View: Introducing ProAlanase, the Newest Mass-Spec Grade Protease from Promega - August 7, 2020
- Go fISH! Using in situ Hybridization to Search for Expression of a SARS-CoV-2 Viral Entry Protein - July 10, 2020
