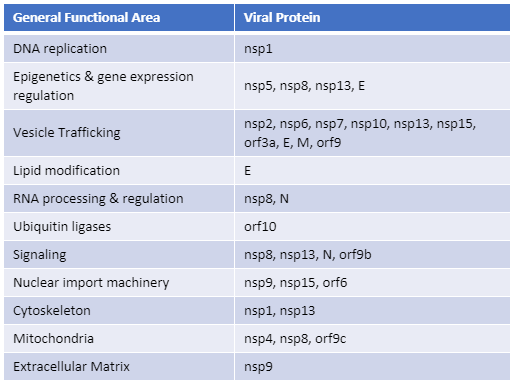
Recently, Gordon et al. published an atlas of protein:protein interactions of all proposed SARS-CoV-2 proteins expressed individually in HEK 293 cells (Table 1). The study tagged each of the viral proteins with an epitope tag and performed a pull-down of the expressed protein followed by trypsin digestion and mass spec analysis, a process referred to as affinity purification–mass spec analysis. The group identified 332 human proteins interacting with 27 SARS-CoV-2 proteins.
The interactions identified in the HEK 293 cells helped Appelberg et al. analyze interactions over time in SARS-CoV-2-infected Huh7 cells. Gordon et al. used the PPI data to identify FDA-approved drugs, drugs in clinical trials, and pre-clinical compounds that bound to the identified human proteins and labs in New York and Paris tested some of these drugs for antiviral effects.
The data from Gordon et al. provides a starting point for detailed analysis of the interactions. Promega offers two systems for live cell measurement of protein:protein interactions (PPI): NanoBRET™ Technology (3) and NanoBiT® Technology (4). Both systems have been cited for investigation of viral:host interactions, however this article will focus primarily on NanoBRET™ Technology.
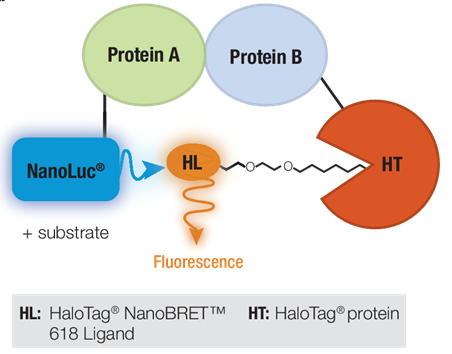
NanoBRET™ Technology is a proximity-based method of measuring PPI through bioluminescent resonance energy transfer or BRET (Figure 1). Two suspected interacting proteins are tagged with either NanoLuc® Luciferase or the self-labeling HaloTag® Protein. The BRET reaction is dependent upon energy transfer from the extremely bright donor NanoLuc® Luciferase to the optimized energy acceptor fluorophore bound to the HaloTag® Protein, HaloTag® NanoBRET™ 618 Ligand. If the two proteins interact, addition of substrate for NanoLuc® Luciferase will produce BRET energy transfer resulting in a fluorescent signal from the NanoBRET™ 618 fluorophore. The ability of the HaloTag® Protein to bind different ligands can be exploited to turn a NanoBRET™ Assay into a pull-down assay (5).
Two recent examples of NanoBRET™ Assays applied to viral:host PPI both concern interactions of phosphorylated viral proteins with host proteins. The hepatitis B virus core protein is needed for capsid formation and requires phosphorylation for function. Nishi et al. used a NanoBRET™ Assay to identify a host phosphatase that could dephosphorylate the core protein. The core protein was expressed as a NanoLuc® Fusion and 150 different human phosphatases were each expressed as HaloTag® Fusions in HEK293 cells. Two phosphatases produced strong BRET signals and conventional pull-down assays demonstrated the strongest interaction with the pyruvate dehydrogenase phosphatase.
Miyakawa et al. report that the interaction of the HIV-2 protein Vpx with host protein SMHD1 is dependent upon phosphorylation of Vpx. The result of the interaction is proteasomal degradation of SMHD1. To identify which cellular kinase is responsible for phosphorylation of Vpx, Miyakawa first performed in vitro experiments and found 30 kinases capable of the task. However, when these kinases were tested in NanoBRET™ Assays in HEK 293 cells, it was evident that it was PIM1 or PIM2 kinase that phosphorylated Vpx.
The NanoBRET™ Assay has also been cited for examination of viral protein interactions. Two recent examples use the NanoBRET™ PPI Assay to investigate interactions within the viral protein responsible for binding and packaging the viral genome into newly assemble viral particles. Lin et al. used the NanoBRET™ Assay to monitor interaction of Ebola nucleoproteins to identify critical amino acids through site-directed mutagenesis. Koma et al. used the same strategy to investigate critical residues for HIV GAG protein interactions through the capsid-domain of the GAG protein.
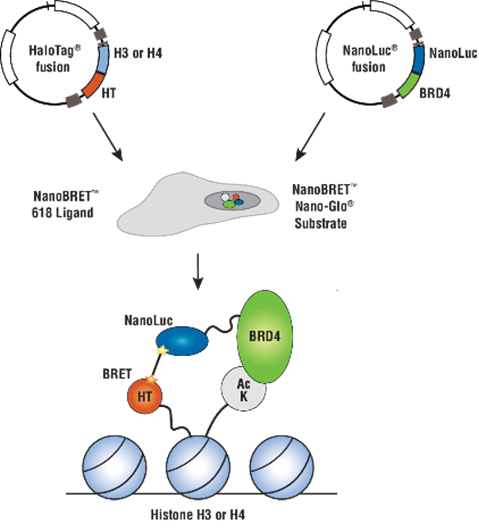
The interactions identified in the Gordon paper immediately suggested NanoBRET™ Assays for further study and their use for antiviral screening. We have many human targets from these studies already available as either HaloTag or NanoLuc fusions via our custom assay service group and these could readily be paired with viral proteins to generate human:viral NanoBRET assays. In addition Promega has pre-designed and optimized assays of several of these same human targets in complex with their native human interactors, for example BRD4 with histone H3.3 (Cat.# N1830; Figure 2). These human:human NanoBRET assays could be used to understand how the presence of viral protein expression may or may not impact these.
Another interesting observation by Gordon et al. was that the viral protein nsp5 interacted with only one host protein, histone deacetylase 2 (HDAC2). The protein nsp5 is a protease, also referred to as 3CL. 3CL is chymotrypsin-like protease and is responsible for releasing many of the nsp proteins from the larger polyprotein made from direct translation of the SARS-CoV-2 RNA genome.
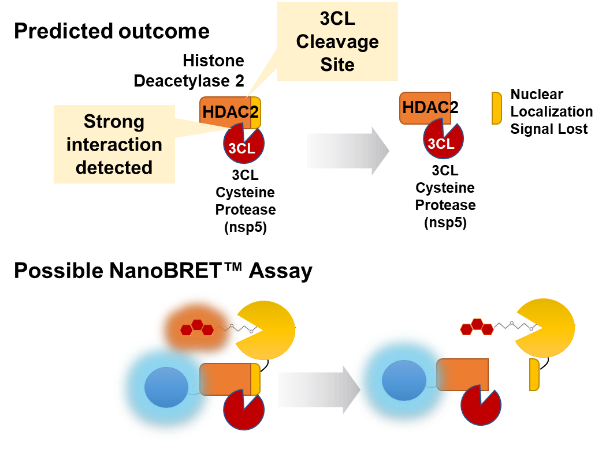
Analysis of the amino acid sequence of HDAC2 revealed a potential 3CL cleavage site near the C-terminus. Curiously, this cleavage site would leave the histone deacetylase domain intact but remove the nuclear localization signal. Cleavage would be expected to prevent translocation to the nucleus where HDAC2 would remove acetyl groups from histones thus affecting transcription. A possible live-cell assay for this would involve placing both NanoLuc® Luciferase and the HaloTag® Protein at opposite ends of the HDAC2 protein. Intact protein would be expected to generate a NanoBRET™ Signal. Cleavage by 3CL would release the NanoBRET™ Pair and would be measured as a decrease in the NanoBRET™ Signal (Figure 3).
Interested in tools and resources for viral research? Visit our web page on SARS-CoV-2 Research, Vaccine, and Therapeutic Development.
References:
- Gordon, D.E. et al. (2020) A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. Nature accelerated article preview. Published online 30 Apr 2020.
- Appelberg, S. et al. (2020) Dysregulation in mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. bioRxiv. Posted 30 Apr 2020.
- Machleidt, T. et al. (2015) NanoBRET-A novel BRET platform for the analysis of protein-protein interactions. ACS Chem. Biol. 10, 1797–804.
- Dixon, A.S. et al. (2016) NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11, 400–8.
- Steffen, L. and Méndez-Johnson, J. (2019) A Luminescent Pull-Down Approach to Confirm NanoBRET™ Protein Interaction Assays.
- Nishi, M. et al. (2020) Prolyl isomerase Pin1 regulates the stability of hepatitis B virus core protein. Front. Cell Dev. Biol. 8, 26.
- Miyakawa, K. et al. (2019) PIM kinases facilitate lentiviral evasion from SMHD1 restriction via Vpx phosphorylation. Nat. Comm. 10, 1844.
- Lin, A.E. et al. (2020) Reporter assays for Ebola virus nucleoprotein oligomerization, virion-like particle budding and minigenome activity reveal the importance of nucleoprotein amino acid position 111. Viruses 12, 105.
- Koma, T. et al. (2019) Allosteric regulation of HIV-1 capsid structure for Gag assembly, virion production, and viral infectivity by a disordered interdomain linker. J. Virol. 93, e00381-19.
