For most molecular biology applications, knowing the amount of nucleic acid present in your purified sample is important. However, one quantitation method might serve better than another, depending on your situation, or you may need to weigh the benefits of a second method to assess the information from the first. Our webinar “To NanoDrop® or Not to NanoDrop®: Choosing the Most Appropriate Method for Nucleic Acid Quantitation” given by Doug Wieczorek, one of our Applications Scientists, discussed three methods for quantitating nucleic acid and outlined their strengths and weaknesses.
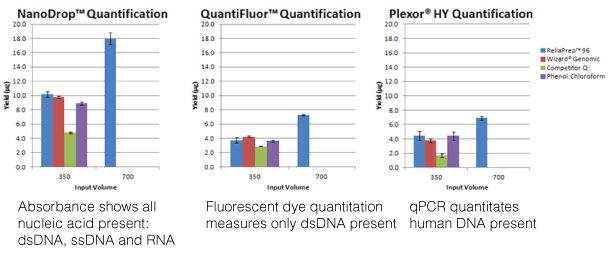
The most commonly used method is measuring the optical density (O.D.) of the nucleic acid solution with the ultraviolet wavelengths 260nm, 280nm, 230nm and 320nm in a spectrophotometer. While this is a simple method for quantitating nucleic acids and assessing the sample’s purity, an O.D. reading is confounded by lack of specificity (all nucleic acids present will absorb at 260nm) and high levels of absorbance at 230nm, indicating contamination, can artificially inflate the 260nm reading and thus, overestimate the amount of nucleic acid present in the sample. This overestimation is especially true with low concentration samples. Unlike when I was in the lab where I had to dilute a portion of my purified nucleic acid into a 1ml cuvette and account for the dilution factor when calculating the concentration, instruments like the NanoDrop® spectrophotometer can measure the concentration directly, using only a 0.5–2µl aliquot of a sample. Nucleic acid concentration can be measured 2ng/µl–15,000ng/µl (15µg/µl), and ratios of A260/A230 >1.7 work well in downstream assays like PCR. However, what if you needed greater sensitivity or needed to assess the fragmentation or degradation of your sample?
Using a fluorescent nucleic acid-binding dye will be more sensitive than spectrophotometry, quantitating down to 50pg in the case of the QuantiFluor® dsDNA System. This sensitivity combined with specificity (e.g., the QuantiFluor® dsDNA System binds only dsDNA) more accurately assesses the nucleic acid you are interested in quantitating. However, to assess concentration, you need to create a series of standards and detect fluorescence on an instrument that uses the wavelengths you need, 504nmEx, 531nmEm for the QuantiFluor® dsDNA System. Furthermore, fluorescent nucleic-acid binding dyes offer no information on purity or integrity. If you use the fluorescent dye method, consider analyzing your sample for integrity using agarose gel electrophoresis.
The final method covered in the webinar was real-time PCR. This method requires the use of PCR reagents and a specialized instrument that can detect the accumulation of the amplification product from the reaction. This adds to the expense of this method but real-time PCR offers sensitivity and specificity as well as higher throughput and the ability to multiplex. However, real-time PCR is also sensitive to PCR inhibitors and nucleic acid fragmentation. One example Doug presented was DNA isolated from FFPE samples, known for degraded nucleic acids. The larger the product you are trying to amplify, the less likely you are to see a PCR product. In this case, designing primers for a smaller amplicon (e.g., 200 nucleotides or less) would increase the chance that the PCR would succeed. Furthermore, real-time PCR offers specificity if the nucleic acid sample comes from a complex sample like saliva, which can contain DNA from human and bacterial cells. If you want to quantitate the human DNA and exclude any contribution from bacterial DNA, using human-specific amplification assay like the Plexor® HY System offers you that specificity.
Depending on your situation, there is a quantitation method that will satisfy your needs. A single method is not perfect but as long as you understand each method’s limitations, you can successfully quantitate your sample while knowing what your chosen method can and cannot tell you.
Sara Klink
Latest posts by Sara Klink (see all)
- A One-Two Punch to Knock Out HIV - September 28, 2021
- Toxicity Studies in Organoid Models: Developing an Alternative to Animal Testing - June 10, 2021
- Herd Immunity: What the Flock Are You Talking About? - May 10, 2021