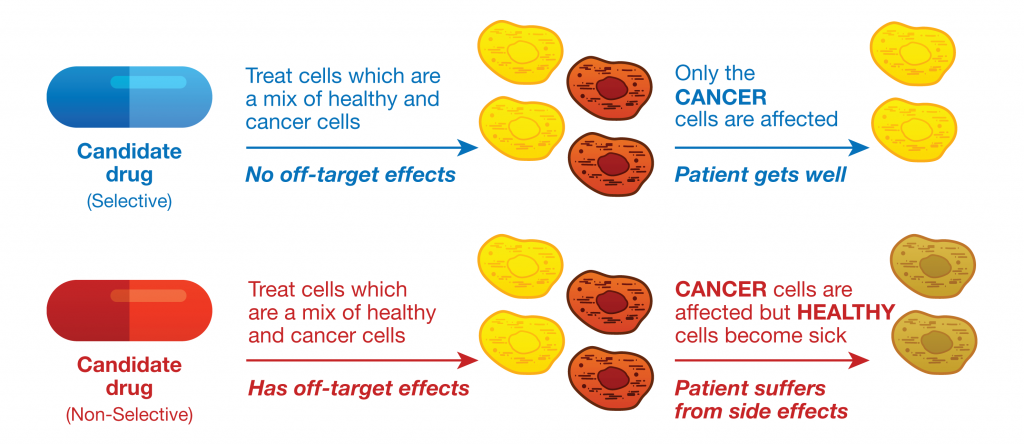
 Drug research and development is a complex and expensive process that begins with initial screening steps of candidate chemical compounds, and compounds that appear to have the desired potency against a specific cellular target or pathway are further evaluated. Candidate compounds that fail late in development or during clinical trials because of off-target effects are costly, and can be dangerous. Therefore drug developers not only need to ensure that a candidate compound is effective as a therapy, but also they need to predict any potential undesirable side effects due to off-target activities as early as possible in the drug discovery and development process.
Drug research and development is a complex and expensive process that begins with initial screening steps of candidate chemical compounds, and compounds that appear to have the desired potency against a specific cellular target or pathway are further evaluated. Candidate compounds that fail late in development or during clinical trials because of off-target effects are costly, and can be dangerous. Therefore drug developers not only need to ensure that a candidate compound is effective as a therapy, but also they need to predict any potential undesirable side effects due to off-target activities as early as possible in the drug discovery and development process.
Cells carry out specific activities such as cell division, growth, and energy usage in response to signals from the environment, to internal signals like DNA damage and even to signals from other cells. These signals are perpetuated and amplified within the cell through biochemical events that modify cellular proteins. One of the most common events of cell signaling is the addition of phosphate groups to proteins (phosphorylation) by kinase enzymes.
Because kinases are one of the largest enzyme classes in the cell and they are involved in many critical cellular processes such as growth, energy use, and cell division, any disruption in their structure or activity leads to diseases such as cancer, inflammation or diabetes. Therefore, kinases are often targeted in drug development efforts, and many small-molecule inhibitors of kinases have already been developed for treatment of different cancers. However, many candidate compounds that show potential also often have off-target effects that make them intractable for drug development. In part this is because molecules that inhibit kinase activity tend to affect the highly conserved regions of these abundant proteins.
Developing methods for profiling the effect of a target compound on the kinase activity (selectivity assessment) is challenging, and many of the current methods are cumbersome and still rely on radioisotopes. First, to profile the selectivity of a compound, many kinases must be assayed at the same time, and one kinase assay method may not be appropriate for all of the kinases being assayed. Second, developing and optimizing assays for each kinase in the selectivity profiling panel and maintaining the stability and supply of the enzyme for the assay is difficult.
 The Kinase Selectivity Profiling Systems are preconfigured and standardized kinase profiling system that can be implemented as a cost-effective in-house screening assay which can be performed by any scientist early in the evaluation of candidate compounds. These systems use a universal kinase assay chemistry that can reliably report on kinase activity regardless of the nature of the substrate because it reports on the production of the universal kinase reaction product, ADP.
The Kinase Selectivity Profiling Systems are preconfigured and standardized kinase profiling system that can be implemented as a cost-effective in-house screening assay which can be performed by any scientist early in the evaluation of candidate compounds. These systems use a universal kinase assay chemistry that can reliably report on kinase activity regardless of the nature of the substrate because it reports on the production of the universal kinase reaction product, ADP.
Such an assay allows drug developers to screen candidate compounds against a wide range of kinases early in the drug development process, potentially identifying problematic off-target effects. In fact this system has been used to describe previously unreported off-target effects for two kinase inhibitors (1). Additionally the same assay can be used to gain more information about the potency of a compound toward a particular targeted kinase or family of kinases later in the screening.
Drug discovery is complex and expensive, and being able to learn as much as possible about candidate compounds early in the screening process is critical to streamlining the process and ensuring that only the most viable compounds progress to later stages of drug development. The flexible bioluminescent kinase strips that we describe here allow researchers to understand more about their target compounds at an earlier stage than ever before.
Literature Cited
Hennek, J. et al. (2016) Bioluminescent kinase strips: A novel approach to targeted and flexible kinase inhibitor profiling. Anal. Biochem. 495, 9–20.
