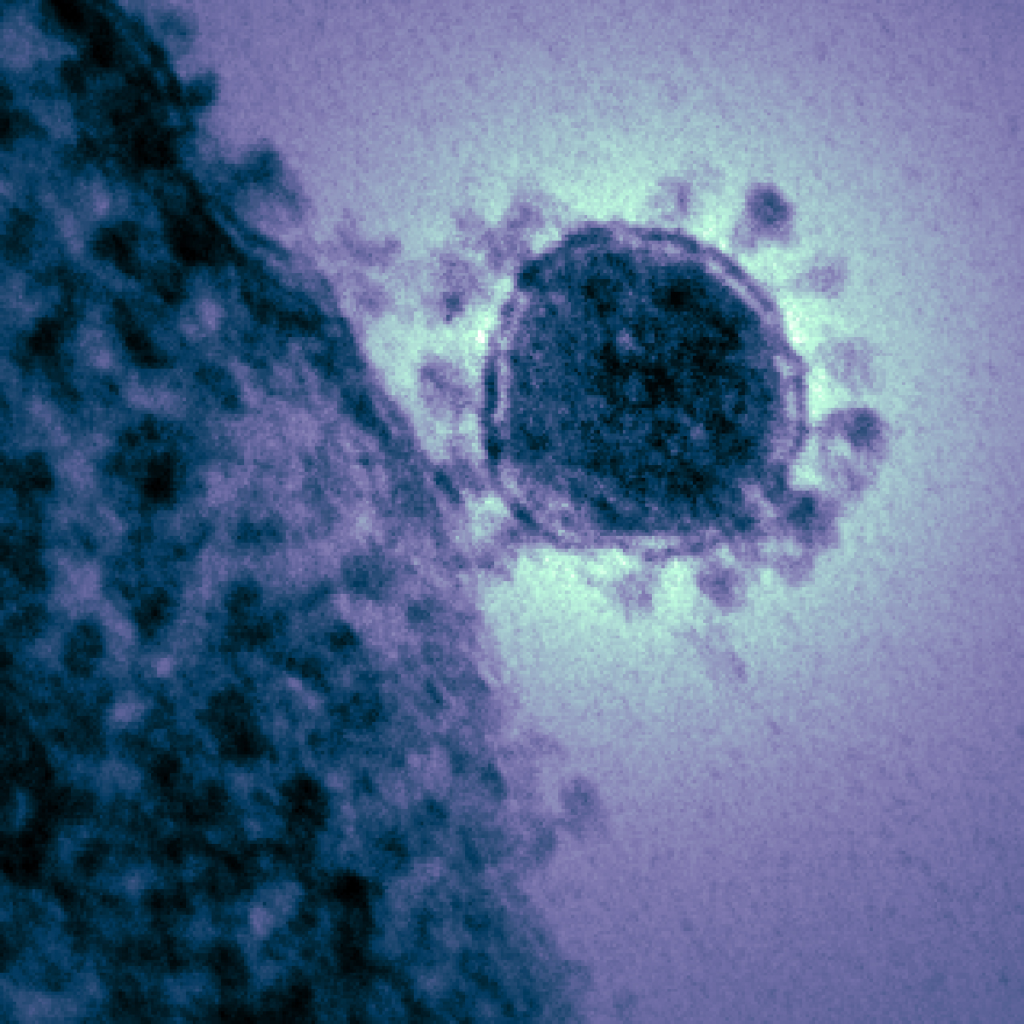
In December 2019, a new disease emerged from a seafood market in Wuhan, China. People who were infected began experiencing fever, dry cough, muscle aches and shortness of breath. The disease swept through China like wildfire and quickly spread overseas to almost every continent. We now know the virus that caused this disease, SARS-CoV-2, is a member of the severe acute respiratory syndrome coronavirus, and the disease itself was officially named COVID-19. According to the Johns Hopkins University Coronavirus Resource Center, there are 877,422 confirmed cases of COVID-19 worldwide, and 43,537 total deaths at the publication of this blog. Those numbers are only expected to increase over the next few weeks.
In this moment of crisis, scientists all around the world are desperately trying to find ways to treat and prevent the disease. One strategy for preventing the spread of the virus is to block its entry into human cells. But first we need to understand how SARS-CoV-2 enters human cells. A research group at the German Primate Center led by Dr. Stefan Pohlmann provides some answers in a recent publication in Cell.
Dr. Pohlmann’s group began by comparing the viral sequences of SARS-CoV-2 and its predecessor—SARS-CoV. SARS-CoV originated from bats and emerged in humans as a severe respiratory disease in 2002. It caused a global pandemic which was finally stopped in 2003. Imagine a Styrofoam ball with ball pins stuck all around it. That’s kind of what SARS-CoV looks like under an electronic microscope. The ball pin-like structures are called spike protein (SARS-S), and the surface unit (S1; the “ball” of the ball pin) is what binds to a cellular receptor (ACE2) that resides on the surface of human cells. Once the S1 subunit is bound to ACE2, cellular proteases (TMPRSS2 and CatB/L, both residing on the surface of human cells) cleave the S protein, allowing another spike protein subunit (SARS-S2) to fuse the viral and cellular membranes. As the membranes fuse, the virus shell is broken apart, releasing RNA into the host cell where it replicates and generates more viral particles.
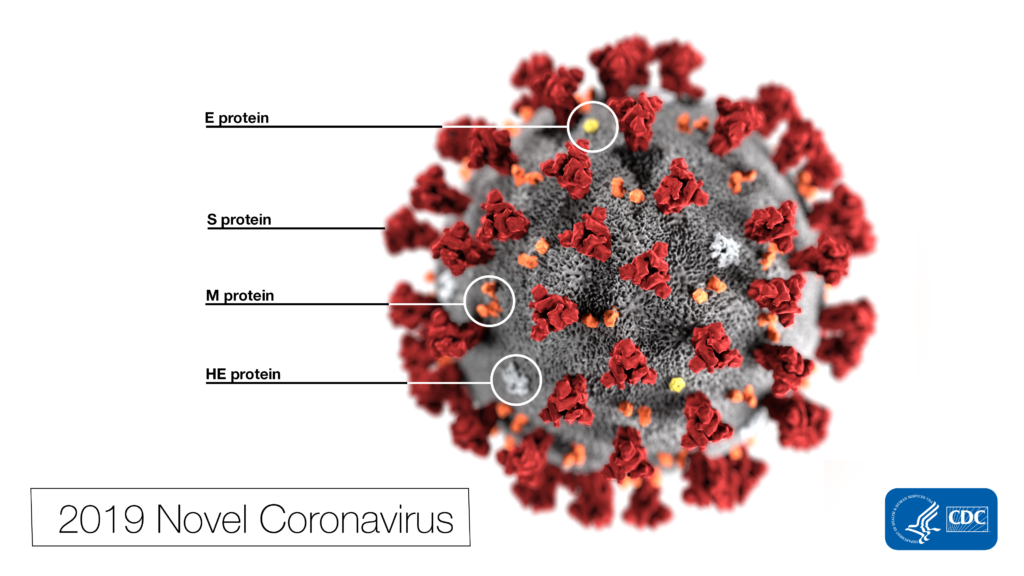
SARS-CoV-2 and SARS-CoV are very similar. In fact, the spike protein of SARS-CoV-2 (SARS-2-S) is 76% identical with SARS-S. Does this mean they enter host cells the same way? The authors set out to test this hypothesis. First, they showed that ACE2 is required for SARS-CoV-2 cell entry. This was supported by the following evidence: 1) Specific amino acids essential for ACE2 binding in SARS-S were also present in SARS-S-2, 2) Overexpression of ACE2 in cells drove more SARS-2-S to enter cells, and 3) An antiserum known to block ACE2 receptors prevented SARS-2-S entry.
Next, they wanted to know whether SARS-CoV-2 depends on the same cellular proteases as SARS-CoV for cell entry. The answer is yes. They found that SARS-2-S uses both TMPRSS2 and CatB/L to enter cells, similar to SARS-S. In addition, treatment with a TMPRSS2 inhibitor (camostat mesylate) blocked the entry of SARS-CoV-2 into primary human lung cells. This is great news as camostat mesylate is already approved in Japan for human use, although for an unrelated illness (pancreatic inflammation). There is hope that this compound, or a modified version, may be used for off-label treatment in COVID-19 patients.
Another hopeful result from this study was that antibodies against SARS-CoV also block entry of SARS-CoV-2—although less efficiently. This means that antibodies generated from SARS-CoV infection or vaccination may also provide some level of protection against SARS-CoV-2.
This study provided important insights into how SARS-CoV-2 infects and enters host cells, and potential targets for antiviral treatment. To combat this global pandemic, we need more studies like this. Rest assured, there are thousands of scientists working tirelessly trying to find out more about how SARS-CoV-2 works, and how we can treat or prevent this disease (if you are one of them—thank you!).
Featured product:
The CellTiter-Glo® Luminescent Cell Viability Assay was used in this study to determine cell viability after camostat mesylate treatment. See more publications demonstrating how CellTiter-Glo was used to detect cytotoxicity of viruses, including high-throughput applications:
SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death
Related Posts
Latest posts by Johanna Lee (see all)
- How Avian Influenza Crosses Species - February 22, 2024
- Macrophages: The Unsung Heroes of Immune Response and Biologic Drug Development - December 28, 2023
- Treating Solid Tumors: Combining CAR-T Cell Therapy with Probiotics - October 31, 2023
