Most, if not all, processes within a cell involve protein-protein interactions, and researchers are always looking for better tools to investigate and monitor these interactions. One such tool is the protein complementation assay (PCA). PCAs use a reporter, like a luciferase or fluorescent protein, separated into two parts (A and B) that form an active reporter (AB) when brought together. Each part of the split reporter is attached to one of a pair of proteins (X and Y) forming X-A and Y-B. If X and Y interact, A and B are brought together to form the active enzyme (AB), creating a luminescent or fluorescent signal that can be measured. The readout from the PCA assay can help identify conditions or factors that drive the interaction together or apart.
A key consideration when splitting a reporter is to find a site that will allow the two parts to reform into an active enzyme, but not be so strongly attracted to each other that they self-associate and cause a signal, even in the absence of interaction between the primary proteins X and Y. This blog will briefly describe how NanoLuc® Luciferase was separated into large and small fragments (LgBiT and SmBiT) that were individually optimized to create the NanoBiT® Assay and show how the design assists in monitoring protein-protein interactions.
Splitting NanoLuc® Luciferase and the Creation of LgBiT
To create a protein complementation assay based on NanoLuc, a suitable splice site was found 13 amino acids from the C-terminus, separating the NanoLuc enzyme into small (amino acids 157–169) and large fragments (amino acids 1–156). The large fragment did not express well and was unstable. To solve this problem, the fragment was mutagenized; 16 individual mutations resulted in a stable, easily expressed fragment that could interact with the 13 amino acid C-terminal peptide to form an active luciferase. The result is the fragment LgBiT, which is soluble in E. coli and expresses well in mammalian cells. The details of the 91 splice sites investigated and the effort to create LgBiT are described in Dixon et al., published in ACS Chem Biol in November 2015.
Designing SmBiT for Low Affinity LgBiT Interactions
LgBiT and the native 13 amino acid peptide, or NP, exhibit an apparent KD of 900nM, an affinity too high for use with weakly interacting proteins. 350 variants of the NP sequence were tested and peptides identified that spanned 5 orders of magnitude in affinity toward LgBiT. One peptide, 114, was only 11 amino acids long with three amino acid substitutions and had an apparent KD of 190µM and was dubbed SmBiT. (By the way, a peptide with greater affinity (KD = 700pM) than the NP was identified. You should be hearing about HiBiT soon!) Again, see Dixon et al for complete details.
Why Minimize the Affinity of the Interacting Partners?
You do not want the LgBiT and SmBiT pair driving the interaction of the two proteins, instead you want the interaction of the two proteins of interest to drive the interaction of the NanoBiT pair. A lab in Belgium definitely understood how unwanted interaction between two parts of a split reporter can be a problem. Their experiments with both split Renilla luciferase and split Venus fluorescent protein suffered from low signal-to-background ratios because of the propensity of the two reporter fragments to self-associate.
Brightness Means Less Is More
The NanoBiT™ pair must have an affinity for one another and, as you would expect, proximity is a key factor in driving these two together. Overexpression of NanoBiT, or any complementation system for that matter, could increase the local concentration of the complementary pairs, driving them together, increasing background and creating a false-positive result. The brightness of NanoLuc luciferase gives the NanoBiT assay an advantage here.
Because NanoLuc® luciferase is 100X brighter than firefly or Renilla luciferase, you need 100-fold less NanoLuc to get a signal equivalent to that of firefly or Renilla luciferases. Likewise, NanoBiT protein interaction assays are much brighter than split firefly luciferase assays at room temperature (21°C) and 37°C. The performance at 37°C is also attributed to the increased stability of the LgBiT fragment. Split firefly luciferase assays work much better at room temperature than at 37°C.
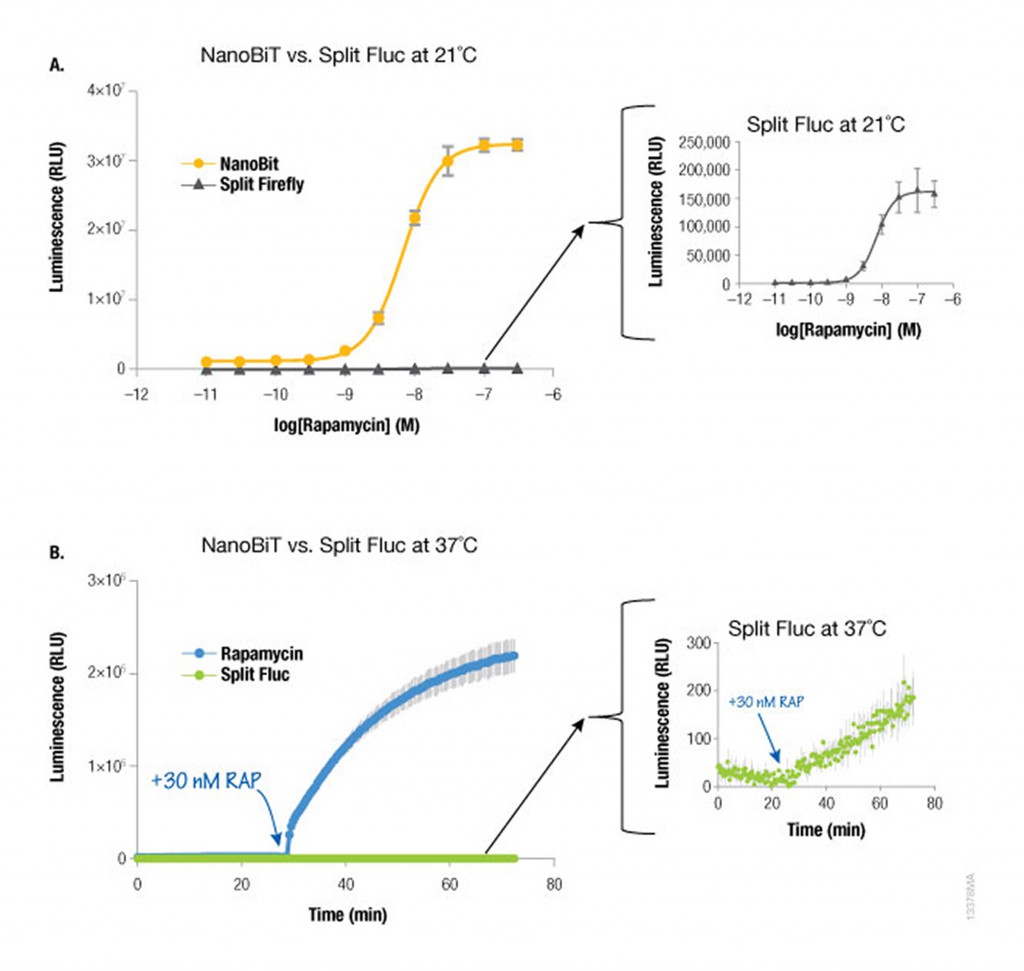
Since NanoBiT is so bright, less protein is necessary to measure an interaction and, thus, you do not need a strong promoter like CMV to drive expression. Consequently, the LgBiT and SmBiT fusion vectors are driven by the TK promoter for low level expression in most mammalian cells, creating assays that are more likely to be reflective of true cellular physiology.
The Take-Home Message
NanoLuc Luciferase was split into two fragments. These fragments were individually optimized into LgBiT and SmBiT to improve expression and lower mutual affinity. The affinity is such that the positive signals from NanoBiT are likely real, and not due to the reporter fragments driving the interaction. The signal from NanoBiT is bright and, requires less protein to generate a readable signal. Taken together, these optimized attributes of the NanoBiT assays allow researchers to monitor protein interactions in live cells with a sensitivity and accuracy that has not been possible before.

