Model organisms are essential tools in the pursuit of understanding biological processes, elucidating the mechanisms of diseases, and developing potential treatments and therapies. Use of these organisms in scientific research has paved the way for groundbreaking discoveries across various fields of biology. In particular, non-mammalian models can be valuable due to characteristics such as rapid life cycles, low cost, and amenability to use with advanced genetic tools, including bioluminescent reporters such as NanoLuc® Luciferase.
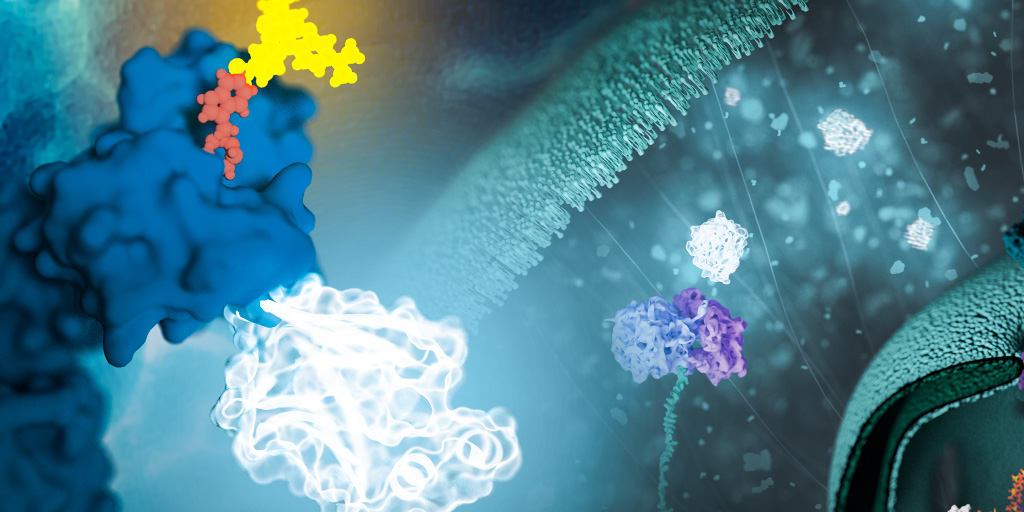
NanoLuc® is a small (19.1 kDa) luciferase enzyme originating from deep sea shrimp that is 100x brighter than firefly or Renilla luciferase. It utilizes a furimazine substrate to produce its bright glow-type luminescence. In the decade following its development, the NanoLuc® toolbox has expanded to include NanoBiT® complementation, NanoBRET™ energy transfer methods, and new reagents such as the Nano-Glo® Fluorofurimazine In Vivo Substrate (FFz) which was designed for in vivo detection of NanoLuc® Luciferase, NanoLuc® fusion proteins or reconstituted NanoBiT® Luciferase. In addition to the aqueous-soluble reagents increased substrate bioavailability in vivo, with fluorofurimazine, NanoLuc® and firefly luciferase can be used together in dual-luciferase molecular imaging studies.
Here we spotlight some recent research that demonstrates how the expanded NanoLuc® toolbox can be adapted to use in non-mammalian models, shedding new light on fundamental biological processes and advancing our understanding of complex mechanisms in these diverse organisms.
Continue reading “Glowing Testimonies: A Review of NanoLuc® Use in Model Organisms”
 Although previous references have provided data regarding the potential oncogenic role of the gene ETV7, there has been minimal investigation as to its physiological role.
Although previous references have provided data regarding the potential oncogenic role of the gene ETV7, there has been minimal investigation as to its physiological role.